Units 4-6 Bio Skibness
1/65
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
66 Terms
calorie
Amount of energy needed to raise the temperature of 1g of Water by 1°C
Calorie
= 1 kilocalorie (i.e. 1000 calories), Amount of energy needed to raise the temperature of 1kg of H2O by 1°C
Calories in Food
1 gram carb = 4 kcal
1 gram protein = 4 kcal
1 gram fat = 9 kcal
1 gram alcohol = 7 kcal
Cellular Respiration
Food energy is converted into chemical energy (ATP) through this process
ATP energy can be formed by breaking carbon–hydrogen bonds.
Energy can be released from molecules that have C–H bonds, such as methane (natural gas), or glucose, or fatty acids.
C6H12O6 + 6 O2 —> 6 CO2 + 6 H2O + 36 ATP
(glucose + oxygen → carbon dioxide + water + energy)
ATP adenosine triphosphate.
ATP is the primary energy molecule in all cells.
• ATP is classified as a nucleoside triphosphate composed of the base adenine, the sugar ribose, and three phosphate groups.
Redox Reactions
Oxidation, Reduction
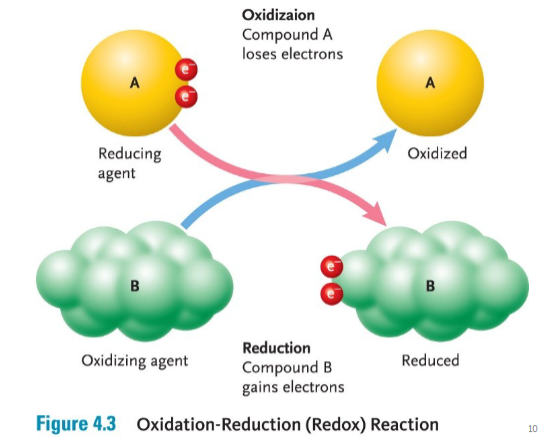
OIL RIG
Oxidation is the loss of electrons, and Reduction is the gain of electrons
During cellular respiration, electrons and hydrogen ions are moved from one molecule to another.
Aerobic respiration
requires oxygen and glucose —> reaction that requires oxygen
Anaerobic respiration
Does not require oxygen
Glycolysis (Cellular Respiration Stage 1) anaerobic
All organisms are capable of doing glycolysis
occurs in the cytoplasm
does not require oxygen to generate ATP
Breaks glucose molecules into two pyruvate molecules
It is the main energy source of prokaryotic organisms that do not have mitochondria.
Glycolysis is a sequence of chemical reactions that break one glucose molecule (six carbons) into two pyruvate molecules (has 3 carbons).
The breaking of bonds is used to form ATP – called substrate-level phosphorylation.
Phosphorylation is the process of adding a phosphate to ADP.
NET GAIN OF 2 ATP (small amount yielded)
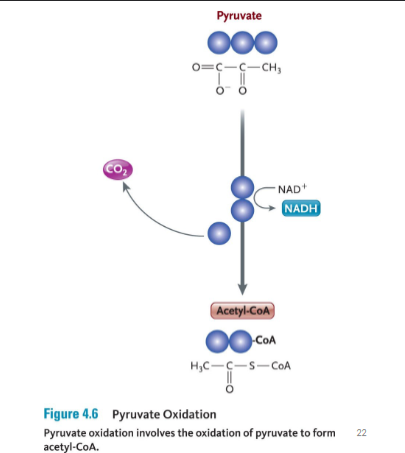
Pyruvate Oxidation (Post glycolysis) aerobic
Oxygen present
occurs in mitochondrial matrix
pyruvate molecules diffuse through mitochondrial membranes into the matrix
one carbon is removed from each pyruvate molecules
When pyruvate is oxidized, a single of its three carbons is cleaved off by the enzyme pyruvate dehydrogenase.
This carbon leaves as part of a CO2 molecule.
In addition, a hydrogen and electrons are removed from pyruvate and donated to NAD+ to form NADH
The remaining two-carbon fragment of pyruvate is joined to a cofactor called coenzyme A (CoA), which is formed from vit B5
The final compound is called acetyl-CoA.
No ATP are made in this step
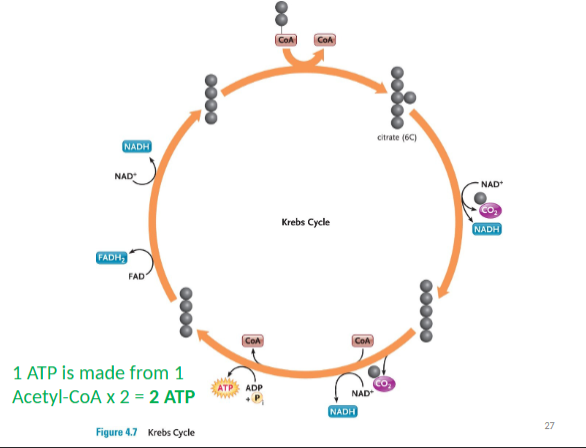
Krebs Cycle
2 cycles per Acetyl-CoA
occurs in mitochondrial matrix
Acetyl-coA combines with oxaloacetate to form citrate
(citric acid – therefore also called citric acid cycle)
harvests energy-rich electrons and protons (NADH) through a cycle of oxidation reactions
electrons passed to an electron transport chain
Acetyl-CoA enters the cycle and binds to a four-carbon oxaloacetate molecule, forming a six-carbon citrate (citric acid) molecule.
Two carbons are removed as CO2 and their electrons donated to NAD+. In addition, an ATP is produced.
The four-carbon oxaloacetate molecule is recycled, and more electrons are extracted, forming NADH and FADH2.
2 ATP PRODUCED PER Acetyl-CoA
Electron transport chain (ETC) and chemiosmosis
occurs in the cristae (inner membrane folds) of mitochondria
Electrons from NADH and FADH2 power the production of ATP
Remember: NAD+ and FAD are reduced to form NADH and FADH2 during glycolysis, pyruvate oxidation and Krebs cycle – all those produced move are needed in this final step
NADH and FADH2 transfer their hydrogens (also called protons or H+) and electrons to a series of membrane-associated molecules called the ETC
Protons are moved through cristae via proton pump membrane proteins to create a large H+ gradient in the intermembrane space
The last transport protein donates the electrons and hydrogen to oxygen to form water.
It is these electrons which provide the energy needed to pump protos across cristae to create the concentration gradient previously mentioned. (note that while this is active transport, it is not ATP that provides the energy).
The only way the H+ can move back into the mitochondrial matrix is through a specific membrane protein called ATP synthase
Chemiosmosis
ATP Synthase: phosphorylates ADP to form ATP
This happens 32 times! Which = 32 ATP!
Chemiosmosis
process of using a chemical gradient for moving across the membrane, resulting in many ATP molecules
Coenzymes that act as oxidizing/reducing agents
NAD+/NADH
• Nicotinamide Adenine Dinucleotide
• NAD+ is oxidized form; NADH is reduced form
• Coenzyme made from vit B3 (niacin)
FAD/FADH2
• Flavin Adenine Dinucleotide
• FAD is oxidized form; FADH2 is reduced form
• Coenzyme made from vit B2 (riboflavin)
Acetyl-CoA
The remaining two-carbon fragment of pyruvate is joined to a cofactor called coenzyme A (CoA), which is formed from vit B5
The final compound is called acetyl-CoA.
The fate of acetyl-CoA depends on the demands of the cell and the availability of ATP.
If there is insufficient ATP, the acetyl-CoA heads to the Krebs cycle.
If there is plenty of ATP, the acetyl-CoA is diverted to fat synthesis for energy storage.
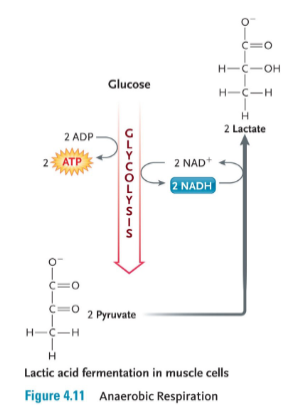
Anaerobic respiration
Requires no oxygen to make ATP
ex. Lactic acid fermentation in mucles
In the absence of oxygen, organisms must rely exclusively on glycolysis to produce ATP.
• But so little is produced that we fatigue very quickly (e.g. we can produce only enough ATP for 20-30 seconds of strenuous exercise)
• Pyruvate from glycolysis instead is turned into the 3-carbon molecule called lactic acid (“lactate”) in a process called fermentation
Bacteria can perform more than a dozen different kinds of fermentation - eukaryotic cells are capable of only a few types of fermentation.
• In yeasts (single-celled fungi), pyruvate is converted into acetaldehyde, which then accepts a hydrogen from NADH, producing NAD+ and ethanol.
DNA Structure
The structure of DNA helps it to function.
The hydrogen bonds of the base pairs can be easily broken to unzip the DNA so that information can be copied.
Each strand of DNA contains the same information (“complimentary” strands),but run in the opposite directions – making them antiparallel.
Having two copies means that the information can be accurately copied and passed to the next generation.
Rosalind Franklin’s work in 1953 using X-ray diffraction revealed that DNA had a regular structure that was shaped like a corkscrew, or helix.
Francis Crick and James Watson elaborated on the discoveries of Franklin and Chargaff and deduced that the structure of DNA was a double helix.
Two strands of DNA bind together by their bases.
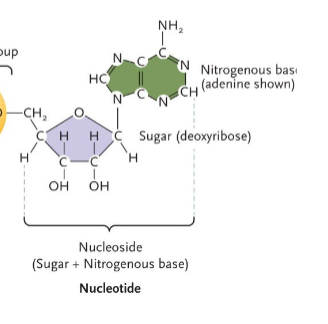
Nucleosides
Our diet is the source of nucleosides in our cells that can be made into nucleotides for DNA replication, or for the transcription of RNA molecules.
• All plant and animal cells contain DNA and RNA (but cannot be processed food).
• We have enzymes in our digestive tract that break down nucleic acids into individual nucleosides that are used to form nucelotides.
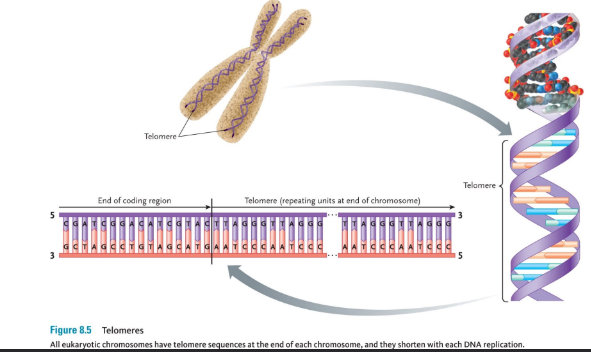
Telomeres
A telomere is a region of repetitive DNA at the end of every eukaryotic chromosome.
• Telomeres protect the end of the chromosome from deterioration (and are 3000-15000 base pairs long).
• A small portion of the telomere s are not replicated during DNA replication and therefore they get shorter after every cell division.
• This is what causes our cells to have a specific lifespan, and this is why we age.
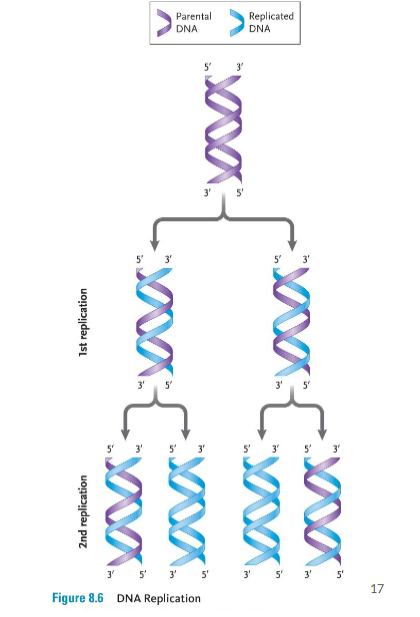
DNA Replication
DNA replicates to a method called “semi-conservative replication – the original strands of DNA separate and act as a template for two new strands
The DNA in all living organisms replicates during the S-phase of the cell cycle (i.e. before mitosis).
In prokaryotes, DNA replication starts at a specific sequence in the genome, the origin of replication
15 - Steps of DNA Replication ):
Replication begins at a point of origin; in eukaryotes there are many points of origin.
An enzyme called helicase unwinds the DNA.
The section where the DNA is unwound is called the replication fork.
Single-strand binding proteins stabilize the separated strands of DNA.
DNA polymerase moves along each strand of unwound DNA and adds the correct complementary nucleotides.
DNA polymerase can only add new nucleotides to an existing strand of DNA.
Therefore, primase adds a small fragment of RNA (an RNA primer) to the initially separated DNA
The RNA primer is complementary to the DNA, and this is later replaced with DNA.
Since the two DNA strands are antiparallel, one strand is oriented as 5’ to 3’, and the other strand is oriented 3’ to 5’.
Polymerase can only add new nucleotides to the 3’ end of the new DNA strand; this is called the leading strand.
The opposite strand is called the lagging strand.
The lagging strand must be replicated in discontinuous segments called Okazaki fragments.
Each segment of the lagging strand must begin with an RNA primer, then polymerase can add nucleotides in the 5’ to 3’ direction. (This is the direction of the new strand being formed.)
Then each RNA primer is removed and replaced with DNA.
DNA ligase is the enzyme that covalently links the new segments of DNA after the RNA primer is removed – forms phosphodiester bonds.
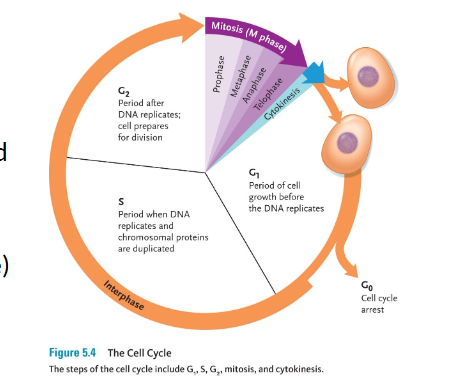
Mitosis
is a cell division mechanism that occurs in somatic cells.
• Mitosis is a continuous process, but it is divided, for ease of study, into four distinct stages:
The cell division that follows interphase is a division of the nuclear contents, known as mitosis.
1. Prophase
2. Metaphase
3. Anaphase
4. Telophase
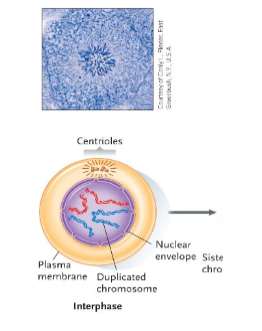
Interphase
Sets the stage for cell division.
Chromosomes are first duplicated.
Although not visible, chromosomes begin to wind up tightly in a process called condensation.
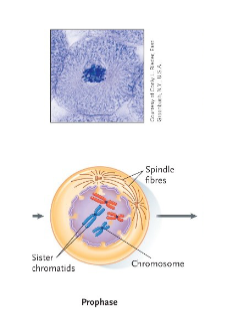
Mitosis – Prophase
Condensed chromosomes first become visible.
The nuclear envelope begins to disintegrate.
The nucleolus disappears.
Centrioles separate in the centre of the cell and migrate to opposite ends (“poles”) of the cell.
The centrioles start to form a network of spindle fibers.
Each cable in the spindle is made of microtubules.
These microtubules grow from each pole until attached to a centromere at a disc of protein called a kinetochore
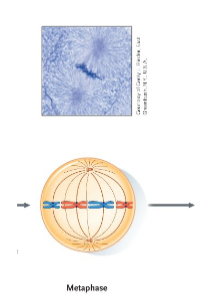
Mitosis – Metaphase
The chromosomes attached to microtubules of the spindle are aligned in the centre of the cell.
• The centromeres are aligned along an imaginary plane that divides the cell in half; this is known as the equatorial plane.
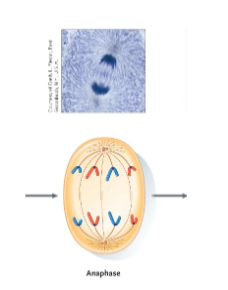
Mitosis – Anaphase
Sister chromatids separate.
• Enzymes break the cohesin and the kinetochores.
• The microtubules of the spindle are dismantled, starting at the poles.
• This pulls the chromatids toward the poles
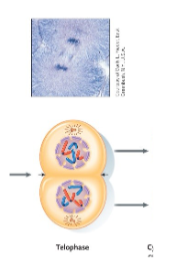
Mitosis – Telophase
The spindle is dismantled.
• A nuclear envelope forms around the set of chromosomes at each pole.
• The chromosomes begin to uncondense.
• The nucleolus reappears.
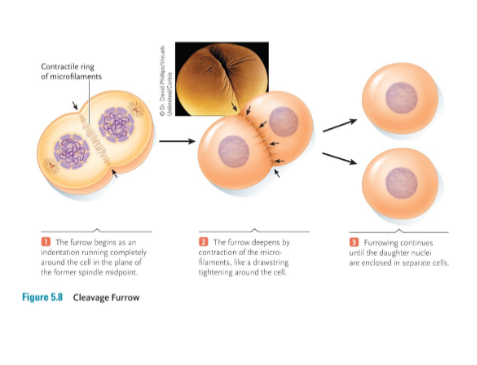
Mitosis – Cytokinesis
Cytokinesis occurs at the end of mitosis and is a division of the cytoplasm into roughly equal halves.
• In animals, cytokinesis occurs by actin filaments contracting and pinching the cell in two.
• This action is evident as a cleavage furrow that appears between the daughter cells.
Cancer
is a growth disorder of cells.
It begins when apparently normal cells grow uncontrollably and spread to other parts of the body.
Benign tumours
are surrounded by a healthy layer of cells (also known as encapsulated) and do not spread to other areas.
Malignant tumours
are invasive and not encapsulated.
• Cells from malignant tumours leave and spread to different areas of the body to form new tumours.
• These cells are called metastases.
Central Dogma of Gene Expression
The information contained in DNA is stored in blocks called genes.
• The strand of DNA that codes for the gene is called the coding strand (sense strand) and the complementary strand is the template
strand (antisense strand) 5
Genes code for mRNA, which then codes for proteins – the proteins determine how a cell will function.
• When gene sequences are used by the cell to make protein, this is called gene expression.
• For example, the human insulin gene sequence is 4044 nucleotides on chromosome number 11.
• Insulin signals cells to take up blood sugar
Transcription
is the process by which a messenger RNA (mRNA) is made from a gene within the DNA. DNA ==> RNA
Has 3 stages
Initiation
elongation
termination
Translation
is the process of using the mRNA to direct the production of a protein by a ribosome. RNA ==> Protein
Messenger RNA (mRNA)
is the “message” that was transcribed from the gene and moves onto the ribosome.
Ribosomal RNA (rRNA)
makes up part of the ribosome.
Transfer RNA (tRNA)
transfers the amino acids to the ribosome during translation.
Initiation (Transcription)
A protein called RNA polymerase produces the mRNA copy of DNA during transcription.
It first binds to one strand of the DNA at a site called the promoter and then moves down the DNA molecule and assembles a complementary copy of RNA. This promotor is called the TATA box – it is not transcribed, but just helps the polymerase locate the start of a gene
The first three nucleotides in the mRNA most commonly are AUG – which code for the amino acid methionine). Start Codon
Elongation (Transcription)
RNA polymerase finds the promoter region of a gene.
Transcription factors are proteins that determine which genes should be transcribed in eukaryotes.
RNA polymerase transcribes only in the 5’ to 3’ direction (same as DNA polymerase in DNA Replication).
The strand of DNA that contains the gene sequence is called the template strand; the opposite strand is the coding strand (because it codes for the gene and will have the same sequence as the mRNA).
Termination (Transcription)
Transcription ends when the RNA polymerase reaches a certain nucleotide sequence that causes transcription to stop; this is called the transcription terminator.
When RNA polymerase encounters the termination sequence, a hairpin loop forms in the mRNA, causing the polymerase and the mRNA to dissociate from the DNA.
From the following DNA sequence, figure out the mRNA sequence.
This sequence is just the gene between the start site and the terminator sequence, so don’t forget to label the 5’ and 3’ ends.
(coding) Sense strand – 5’ ATGGCCTATGAATCG 3’
(template) Antisense strand – 3’ TACCGGATACTTAGC 5’
Therefore, the RNA transcript is 5′AUG GCC UAU GAA UCG 3′.
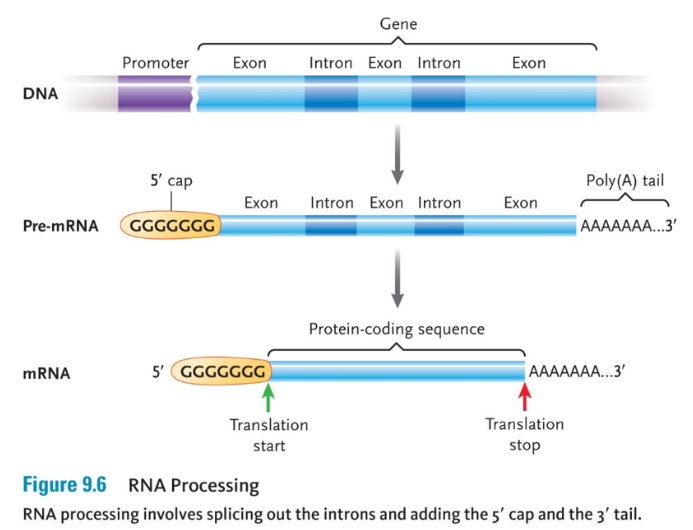
RNA PROCESSING
Exons want to be expressed / Introns removed
RNA PROCESSING
When a eukaryotic cell first transcribes a gene, it produces a primary RNA transcript (also called pre-mRNA) of the entire gene.
Introns do not code for the amino acid sequence and must be removed.
The parts of the gene that code for the mRNA sequence are the exons.
Enzyme-RNA complexes cut out the introns and join together the exons to form a shorter mRNA transcript.
A 5’ cap and a 3’ poly-A tail are also added, which protect the RNA from being degraded.
RNA processing occurs in the nucleus immediately after transcription.
mRNA splicing occurs in a spliceosome, which is a combination of the “pre-mRNA” and small nuclear ribonucleoproteins – snRNPs.
In humans, genes may be spliced together in different ways.
By using different combinations of the same exons, different proteins can be created.
This is termed alternative splicing.
The 25 000 genes of the human genome appear to encode as many as 120 000 different mRNA transcripts. Gene → 5 mrna
Translation Process
Genes are read and translated in the cytoplasm of the cell
To correctly read a gene, a cell must translate the information encoded in the DNA into the language of proteins.
Translation follows rules set out by the genetic code.
The mRNA is “read” in three-nucleotide units called codons.
Each codon corresponds to a particular amino acid.
Translation occurs on ribosomes, which are the protein-making factories of the cell.
Each ribosome is a complex of proteins and several segments of ribosomal RNA (rRNA).
Ribosomes consist of a small subunit and a large subunit
Once an mRNA molecule has bound to the small ribosomal subunit, the other larger ribosomal subunit binds as well, forming a complete ribosome.
During translation, the mRNA threads through the ribosome three nucleotides at a time.
The large subunit contains binding sites for tRNA molecules (they are the A, P, and E sites (Acceptor cite Peptide cite Exit cite)
It is the tRNA molecules that bring amino acids to the ribosome to use in making proteins.
The first tRNA “docks” in the A site of the ribosome
The structure of a tRNA molecule is important to its function:
It holds an amino acid attachment site at one end and a three-nucleotide sequence at the other end.
This three-nucleotide sequence is called the anticodon and is complementary to one of the 64 codons of the genetic code.
Since the first codon of mRNA is AUG, the first amino acid is always methionine (Met)
Before a new tRNA can be added, the previous tRNA in the A site shifts to the P site.
At the P site, peptide bonds from between the incoming amino acid (on the new tRNA in the A site) and the growing chain of amino acids
The original tRNA in the P site eventually shifts to the E site where it is released.
This process needs energy, but GTP is used instead of ATP
Translation continues with tRNA continuously moving along the A, P, E sites, bonding amino acids together, until a stop codon is encountered that signals the end of the protein.
The ribosome then falls apart and the newly made protein is released into the cell.
Multiple ribosomes may transcribe one single mRNA.
Once translation is finished, proteins fold into secondary and tertiary structures
Proteins can be packaged into vesicles that pinch off and move to the Golgi bodies for further processing; this is called post-translational modification.
Within the Golgi bodies, proteins can be combined with lipids or sugars to produce
Lipoproteins (like LDLs made in the liver), or
Glycoproteins (like our blood type markers, A, B, and AB).
Codons
Each codon is made up of three nucleotides.
Each nucleotide in the codon can be one of four nucleotides; therefore, there are (4)(4)(4) = 64 possible codons.
How many amino acids do we have? (20 - 11 - 9essential)
more than one codon can code for the same amino acid as mentioned in the video
Genetic Code
Codons are made fo three nucleotides and make up these codes
Heredity
is the transfer of traits from parent to offspring.
• A heritable feature is called a trait, or phenotype.
• What are some human phenotypes? Eyes, Hair
Alleles
These alleles make up our genotype
Humans have 23 pairs of homologous chromosomes;
therefore, we have two copies of every gene, called
alleles (one from each parent).
Mendel’s Theory of Inheritance
He knew some plants were “true-breeding,” which we now know occurs because the alleles for some traits are the same, that is, homozygous.
• When the alleles for a trait are different, they are heterozygous.
He called the trait expressed in the F1 generation the
dominant trait.
He named the trait not expressed in the F1 generation
the recessive trait.
Punnett Squares
A Punnett square provides a method for determining the possible offspring genotypes and phenotypes from the alleles of each parent
The genotypes of potential offspring are represented within the square.
• Looking at one trait is a monohybrid cross.
Mendel Testcross
used to determine the genotype of unknown individuals in the F2 generation showing a dominant trait
the unknown individual is corssed with a homozygou recessive individual
if the unknown is homozygous dominant, then all of the offspring will express dominant traits.
ff the unknown is heterozygous, then the offspring will express one-half recessive traits and one-half dominant traits.
Non-Mendelian Inheritance – Polygenic
Often the expression of phenotype is not straightforward – i.e. there isn’t always a simple “dominant” versus “recessive” pattern to inheritance.
• Continuous variation. A phenotype (i.e. trait) can show a range of small differences when many genes code for it.
• This type of inheritance is called polygenic. Example: height, skin colour, body weight, intelligence, and different disease states
Non-Mendelian Inheritance - Codominance
A gene may have more than two alleles in a population.
• Sometimes in heterozygotes there is not a dominant allele. Instead, both alleles are expressed.
• These alleles are said to be codominant.
• The gene that determines ABO blood type in humans exhibits more than one dominant allele.
• The gene encodes an enzyme that creates antigens on the RBC cell surface – these act as recognition markers
Ex. Blood types
The gene that encodes the enzyme, designated I, has three alleles: IA,IB, and i.
• Different combinations of the three alleles produce four different phenotypes, or blood-types (A, B, AB, and O).
• Both IA and IB are dominant over i, and also codominant.
Non-Mendelian Inheritance – Epistasis
Epistasis refers to the effects of one gene are modified
by one or more other genes (i.e. modifier genes)
• Epistatic mutations have different effects in
combination than each gene individually.
Ex. Labradors some mutate and come out brown for instance crossing black and white
Non-Mendelian Inheritance – X-Linked
In fruit flies, sex is determined by the number of copies of the X chromosome (same as humans), that an individual possesses.
• A fly with two X chromosomes is female.
• A fly with an X and a Y chromosome is male.
The presence of the white-eyed trait only in males must mean that the white-eye allele occurs only on the X chromosome.
• A trait determined by a gene on the sex chromosome is said to be sex-linked or X-linked.
in humans, some common X-Linked recessive traits
are:
• Colour blindness
• Hemophilia (blood clotting disorder)
• Duchene muscular dystrophy
NOTE: X-linked diseases are much more common in men. Why? men have only one X chromosome, so they cannot compensate for a faulty gene on that chromosome.
Non-Mendelian Inheritance – Linkage
Linkage is defined as the tendency of genes located close together on a chromosome to be inherited together
• The further two genes are from each other on the same chromosome, the less likely they are to be linked, and more likely that they will be separated by crossing over during meiosis
Genetic Disorders – Nondisjunction
Nondisjunction is the failure of chromosomes to separate correctly during either meiosis I or meiosis II.
• This leads to aneuploidy = an abnormal chromosome number.
• Most of these abnormalities cause a failure to develop or an early death before adulthood.
EXCEPTION OF EARLY DEATH WOULD BE TRISOMY 21 (DOWN SYNDROME)
DNA mutations
Because so much DNA is being replicated in the many cells of the body, there is potential for errors to occur,
We have about 130 genes that code for DNA repair enzymes
• Repair enzymes, as well as polymerase itself, compare the new DNA strand with the original DNA strand and fix any incorrect nucleotides.
• This is called proofreading.
• Proofreading is still not perfect and mistakes can occur: i.e. DNA mutations.
DNA mutation types
There are two general ways to alter the genetic message encoded in DNA.
• Point mutations
• Recombination mutations
Point Mutations
There are different types of Point Mutations.
1. Substitution (mismatch) changes the identity of a base or bases.
2. Insertion adds a base or bases.
3. Deletion removes a base of bases.
What could be the consequence of a deletion or addition?
• A frame-shift mutation results.
• These are extremely detrimental because the final protein intended by the message may be altered dramatically, or it may not be made at all.
Ex. THE CAT SAW THE DOG
Substitution: THE BAT SAW THE DOG
Insertion: THE CRA TSA WTH EDO G
Deletion: THE ATS AWT HED OG (the C is missing)
Apoptosis
(programed cell death)
Immune cells that kill cancer cells (natural killer cells)
Mutagen
substance that mutates DNA
Carcinogen
substance that mutates DNA and also causes cancer
How do we acquire DNA mutations
1. Mistakes during DNA replication
2. Transposition
3. Mutagens and carcinogens (most common cause)
4. Viruses
5. Genetic Disorders – i.e. inherited mutations (approximately 10% of diseases are from inherited mutations)
Genetic Disorders
• Phenylketonuria (PKU) – point mutation in liver enzyme gene, which causes brain damage. Babies are tested at birth for presence of enzyme that breaks down phenylalanine into tyrosine (amino acids).
• Nonpolyposis colorectal cancer – autosomal dominant, repeated CA mutation in DNA repair enzyme gene = a non-functional repair protein
• Sickle-cell anemia is an autosomal recessive disease where the hemoglobin gene is mutated, causing abnormal red blood cell formation and reduced capacity to carry oxygen.
• Tay Sachs is an autosomal recessive disease where the HEXA gene that codes for a lysosomal enzyme is mutated, causing brain deterioration (leading to death most often before adolescence).
• Cystic fibrosis is an autosomal recessive disease where the chloride channel gene is mutated, causing altered water balance inside of cells, and this leads to excessive mucus production, which impacts the lungs, liver, pancreas, and sweat glands.
• Huntington’s disease is an autosomal dominant disease where CAG nucleotide repeats are inserted into a gene that increases how many glutamine amino acids are incorporated into the protein. Greater than 35 repeats causes the disease which involves brain deterioration beginning usually in the late 30s.
• Hemophilia is a recessive X-linked disease that affects a gene involved in blood clotting. A lack of any of the blood clotting factors can lead to an inability to stop bleeding.
• Duchene muscular dystrophy is a recessive X- linked disease that involves an important muscle protein, called dystrophin, that holds myofibrils together. Without proper functioning of this protein, muscles degenerate. The disease affects children from early childhood and usually a wheelchair is required by age 10 to 12.
How do our cells deal with mutations?
• Proofreading
Proofreading
Approximately one in every 100 000 to 1 million base pairs will be a mismatch (substitution) during replication.
• Generally, a mismatch causes the polymerase enzyme to pause, and then remove the incorrect base pair using another enzyme function called exonuclease activity; this is proofreading.
• Then the error can be removed, and the correct nucleotides replace the mistake.
• Repair enzymes
Meiosis
Asexual Reproduction – no genetic exchange. Offspring have same DNA as parent.
Sexual Reproduction – genetic variation, gamete production via meiosis
• All eukaryotic organisms have some version of meiosis.
Why does meiosis occur?
• To produce gametes (sperm or eggs)
• Gametes are haploid cells (have half the normal amount of DNA).
What cells undergo meiosis?
• Germ cells in the testis or ovary (in mammals)
What is the result of meiosis in humans?
• Sperm and eggs are the result.
• Sperm fertilize an egg to produce a zygote.
Two unique features not found in mitosis
Meiosis has two unique featuresnot found in mitosis.
1. Synapsis (Prophase I)
• This is the process of drawing together homologous chromosomes down their entire lengths so that crossing over can occur.
2. Reduction division
• Because meiosis involves two nuclear divisions but only one replication of DNA, the final amount of genetic material passed to the gametes is half of the original cell.
Crossing over occurs between two non- sister chromatids of homologous chromosomes.
• The chromatids break in the same place, and a section of chromosomes are swapped.
• The result is a hybrid chromosome.
• The pairing is held together by the cohesin proteins between sister chromatids and the cross-overs.