Chapter 13 Part 2 (slides 20-42)
1/16
Earn XP
Description and Tags
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
17 Terms
basic requirements for translation
mRNA
charged tRNAs
ribosome
initiation factors, elongation factors
energy sources
primers are NOT required
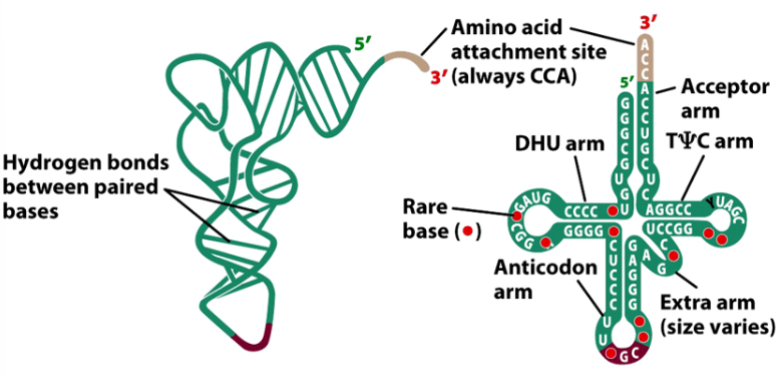
tRNA structure
amino acid is attached to the 3’ end (always CCA)
anticodon loop base-pairs with a codon in mRNA
has hydrogen bonding between paired bases
e.g. cloverleaf model, has different arms (see picture)
unusual bases found in tRNAs
result from post-transcriptional modification of bases in tRNAs
inosine: a modified adenine (know this one)
others (don’t worry about them): inosinic acid (I), 1-methyl inosinic acid (Im), 1-methyl guanylic acid (Gm), NN-dimethyl guanylic acid (Gm_), pseudoridylic acid, ribothymidylic acid (T)
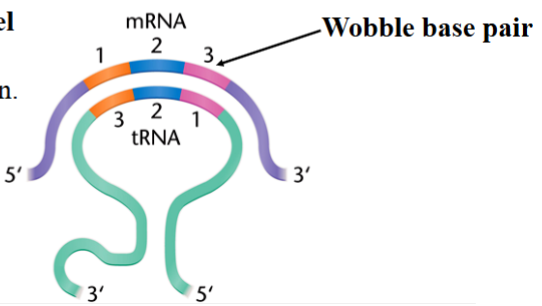
wobble hypothesis
interaction between the third position of the codon in mRNA and the first position of the anticodon in the tRNA is less critical and less constrained
some tRNA bases can pair with multiple mRNA bases at the wobble position
allows translation to occur without the need for the cell to synthesize all 61 tRNAs
e.g. inosine: a post-transcriptionally modified adenine that can occur at the wobble site of tRNA
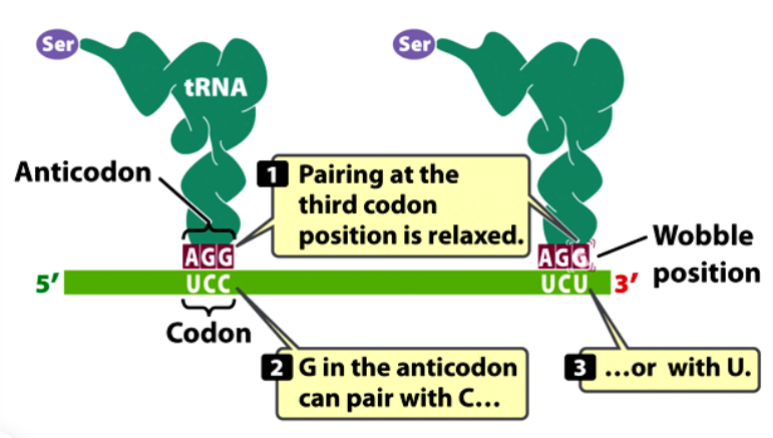
anticodon-codon base pairing rules
(listed as 1st tRNA position (5’) first, 3rd mRNA position (3’) second)
A pairs with U
C pairs with G
G pairs with C or U
U pairs with A or G
I pairs with A, U, or C
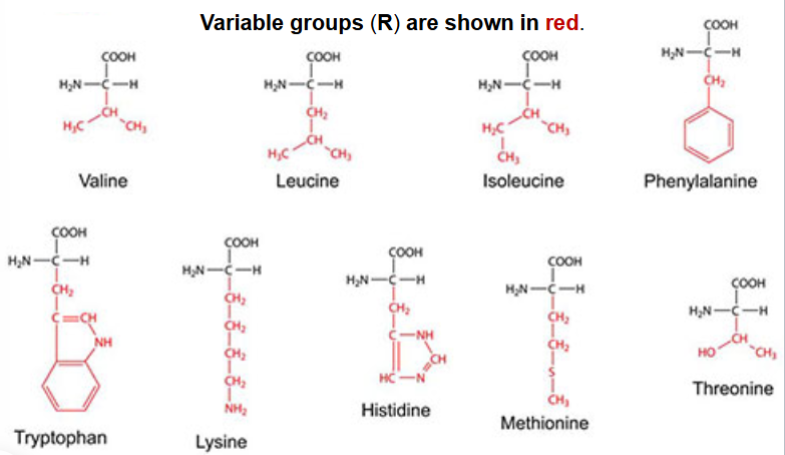
essential amino acids for humans
valine
leucine
isoleucine
phenylalanine
tryptophan
lysine
histidine
methionine
threonine
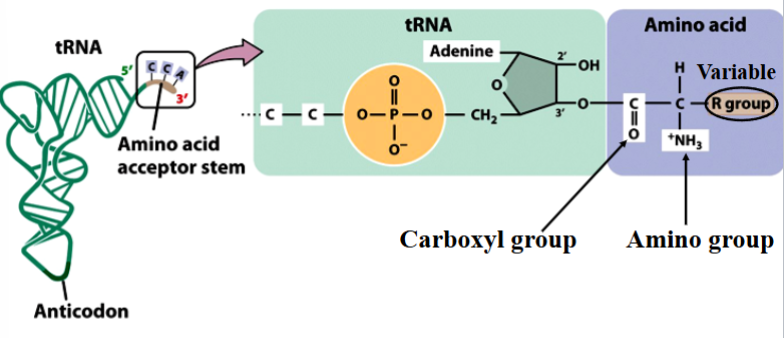
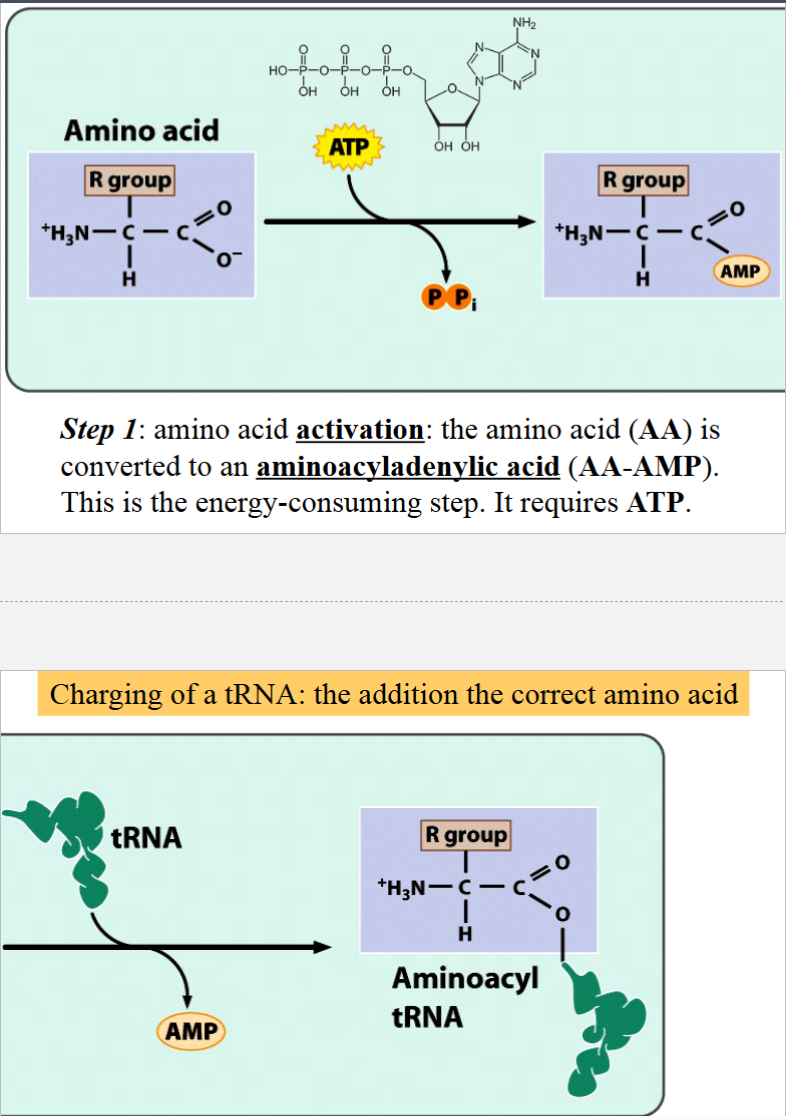
the carboxyl (C=O) of the amino acid is covalently attached to the 3’ end of a tRNA
charging of tRNA
the addition of the correct amino acid
catalyzed by 20 different aminoacyl tRNA synthases
synthases recognize amino acids by their R groups, and recognize tRNAs by their shapes and base sequences
energy is spent in the process, eventually makes a new peptide bond in the ribosome
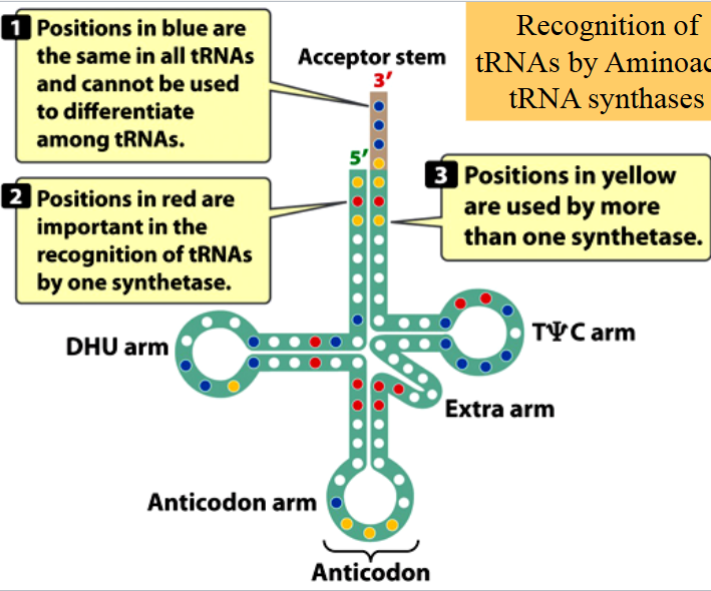
how aminoacyl tRNA synthases recognize tRNAs
some bases are the same in all tRNAs, cannot be used to differentiate among different tRNAS (e.g. ACC at the amino acid attachment site) (blue in the picture)
some bases are important to be recognized by only one synthetase (red in the picture)
some bases are used by more than one synthetase (yellow in the picture)
steps in the charging of a tRNA
step 1: amino acid activation (convert AA to an aminoacyladenylic acid (AA-AMP)
note: step 1 is energy-consuming, requires ATP
step 2: charging (AA-AMP loses AMP, AA is transferred to the 3’ end of a tRNA)
result of step 2: aminoacyl tRNA (charged tRNA)
ribosome
a large particle of rRNA and proteins where translation occurs
made of two subunits: small and large
Svedberg unit
a measure of the rate at which particles sediment in a centrifugal field
NOT a measure of molecular weight
depends on weight, shape, and size
unit (S) can be used for measuring large molecules or large cell components, e.g. ribosomes and organelles
Svedberg values for prokaryotic ribosomes
small subunit: 30S
large subunit: 50S
complete ribosome: 70S
note: S values for small and large subunit do not add up perfectly to the value for the complete ribosome. This is okay.
sites in the ribosome
A: aminoacyl
P: peptydyl
E: exit

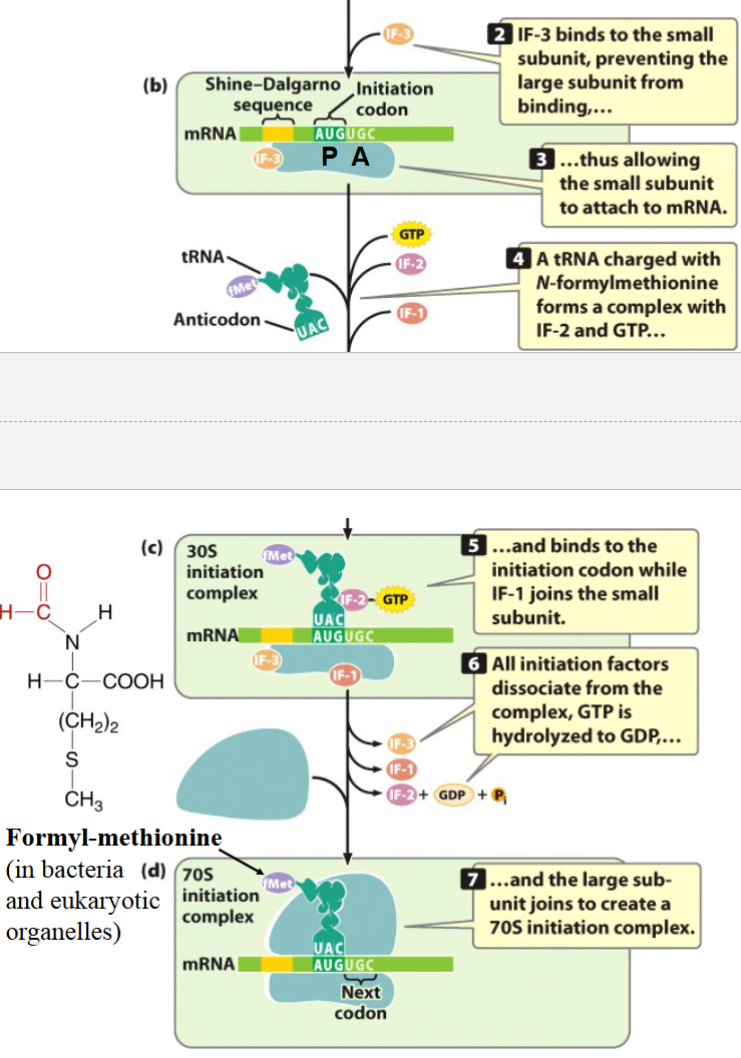
steps in initiation of translation
1) mRNA binds to the small ribosomal subunit with AUG positioned on the P site
2) f-Met-tRNA (in prokaryotes) binds to AUG codon
3) the large ribosomal subunit joins the complex (requires GTP for energy plus IF proteins (initiation factors))
(eg. IF-3 binds to small subunit in step 1 to prevent the large subunit from binding)
see picture for more detail
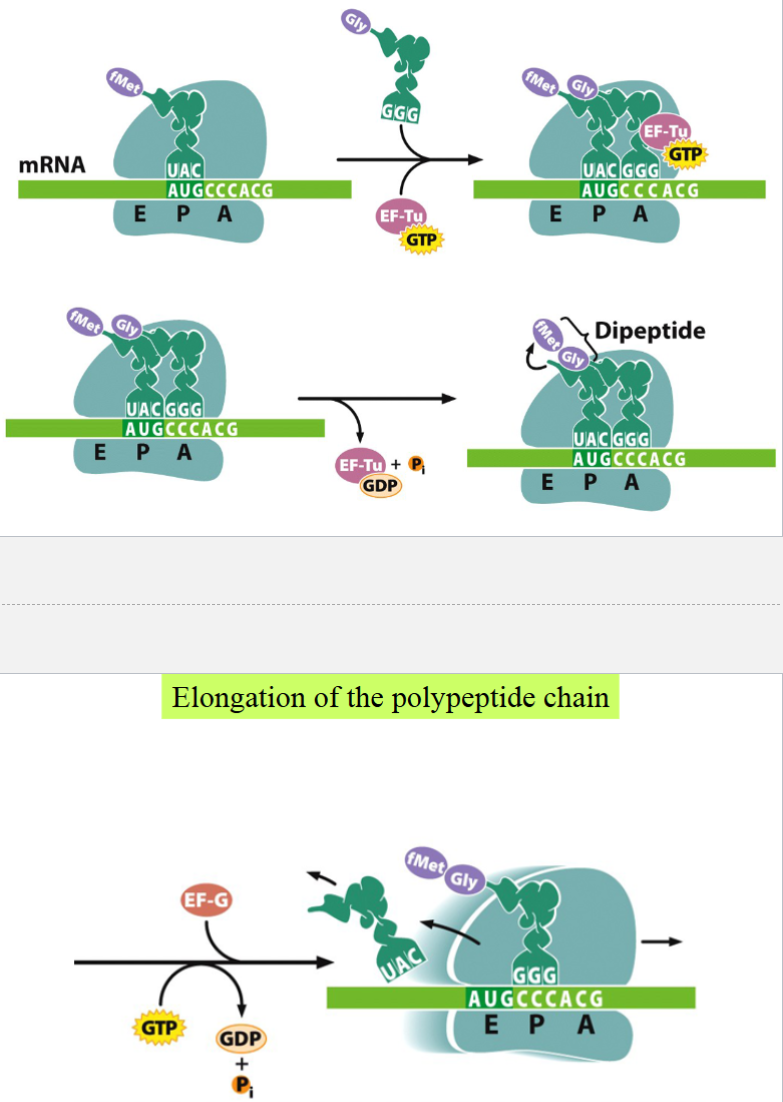
elongation of the polypeptide chain
EF-Tu (an elongation factor) and GTP facilitate the binding of the second tRNA to the second codon at the A site
AA on the first tRNA is transferred, forms a peptide bond with the AA on the second tRNA → forms a dipeptide
first tRNA moves to the E site
mRNA is shifted to place the second codon in the P site and bring the third codon to the A site
tRNA carrying the third AA binds to the third codon on the A site
dipeptide on the second tRNA is transferred → forms tripeptide attached to the third tRNA
second tRNA moves to E site with help of EF-G and GTP
third codon moves to P site
cycle repeats, etc.
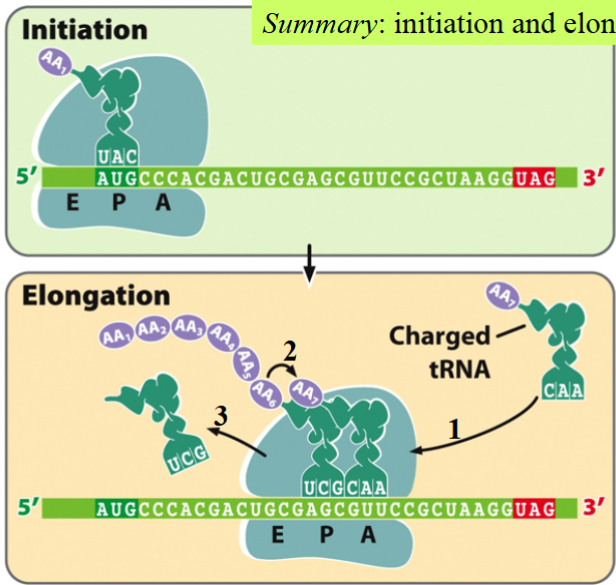
polypeptide chain init9ation and elongation, summarized/condensed
AUG (mRNA) binds at P
UAC (tRNA) attaches to AUG
a new tRNA attaches to the next codon at A
with energy, the amino acids attached to both tRNAs join (remain attached to the second tRNA)
both tRNAs slide over 1 (now UAC is at E, next one is at P)
first tRNA (UAC) pops off
cycle repeats
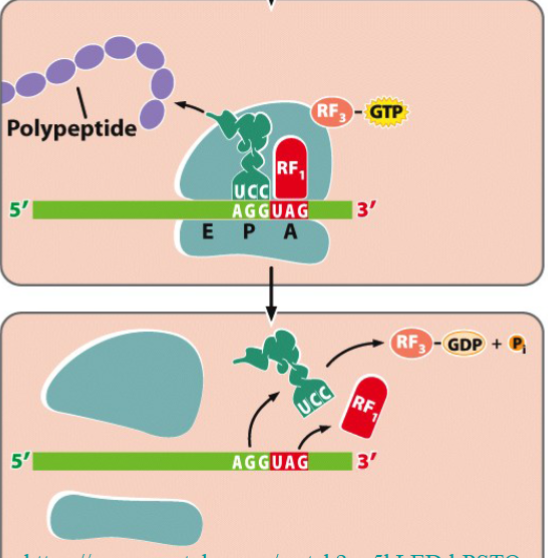
termination of translation
a stop codon moves to the A site of the ribosome
no tRNA binds to the stop codon
RF1 (release factor 1) binds to the stop codon
RF3 (GTP-dependent release factor 3) releases the polypeptide chain from the last tRNA
the entire translation complex dissociates