Nitrogen metabolism
1/19
Earn XP
Description and Tags
18
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
20 Terms
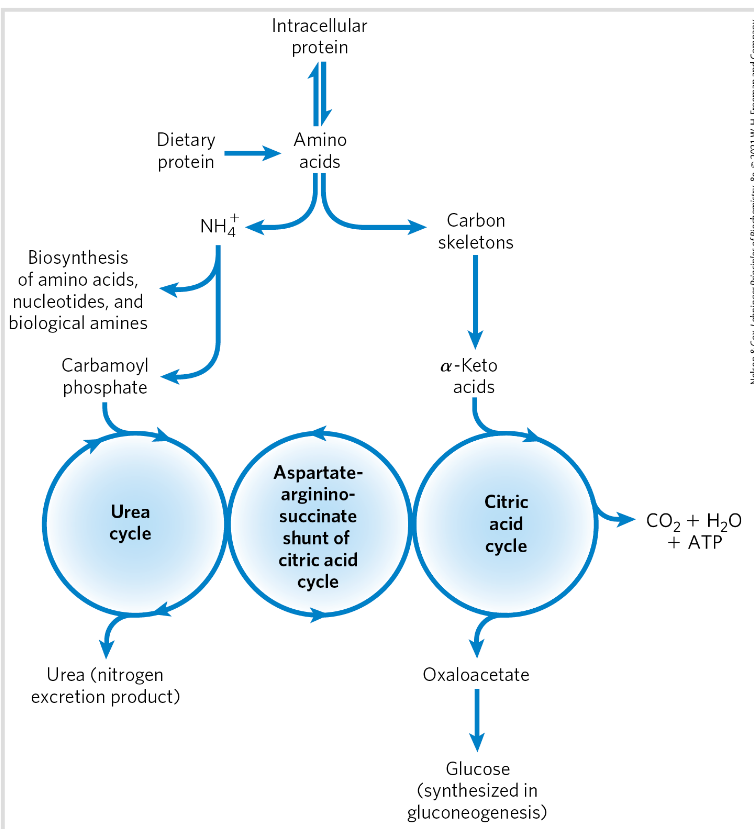
Degradation of amino acids has 2 ways
Either via amino groups or carbon skeletons
NH3→ toxic- has to be excreted in a harmless form
Important precrusor in biosynthesis
When are amino acids degraded?
When normal protein turnover are not needed for new protein synthesis
Ingested amino acids exceed the body’s needs for protein synthesis
Cellular proteins are used as fuel because carbohydrates are not available or during starvation or diabetes mellitus
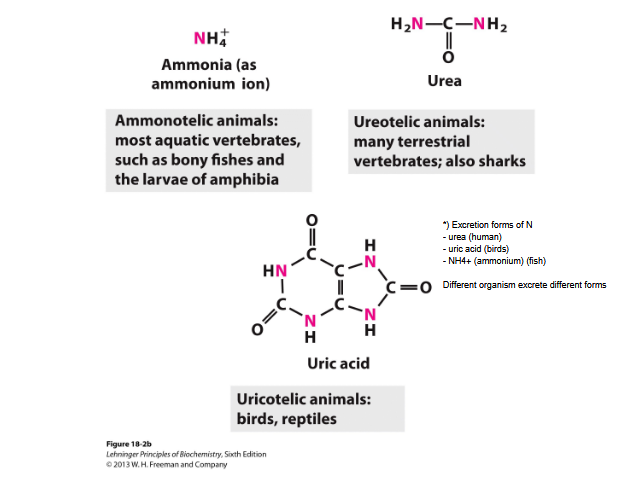
Excretion of amino groups
Fish- excrete amino nitrogen as ammonia
Terrestrial animals - urea
Birds and reptiles- uric acid
Amino acids converted into the citric acid cycle
Glutamate and glutamine - to alfa-ketoglutarate
Alanine→ pyruvate
Aspartate → OAA
Can all be used in citric acid cycle
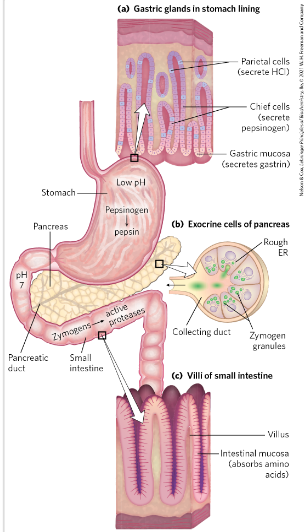
Uptake of amino acids from diet
mechanic degradation of food
- proteases - in stomach and intestine
- uptake over intestinal mucosa -> blood-> liver
Pepsinogen converted to Pepsin=protease-> degrade proteins→ smaller peptides
Low pH-> why? Not healthy for the tissue-> but
pepsin needs to be activated (optimal pH=2
Helps unfold globular proteins and bonds more accessible to enzymatic hydrolysis
When food enters the small intestine→ low pH trigger hormonal release of secretin in the blood→ stimulates pancreas to release bicarbonate→ neutralizes the gastric HCl in small intestine
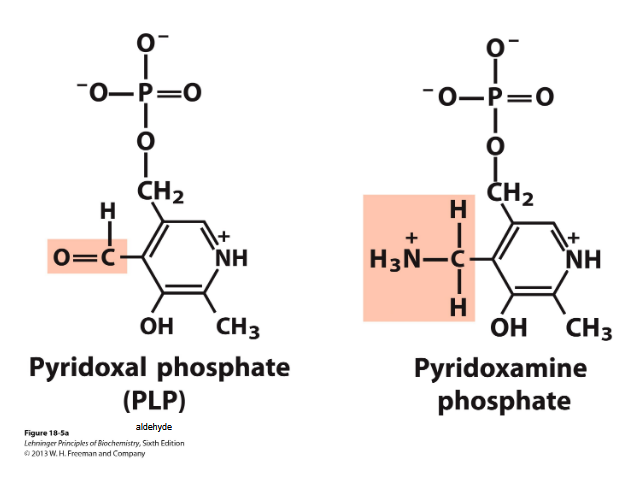
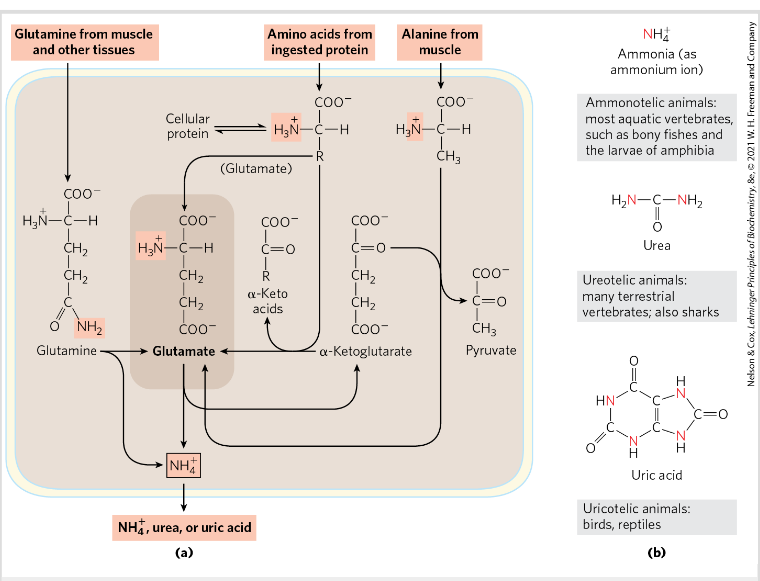
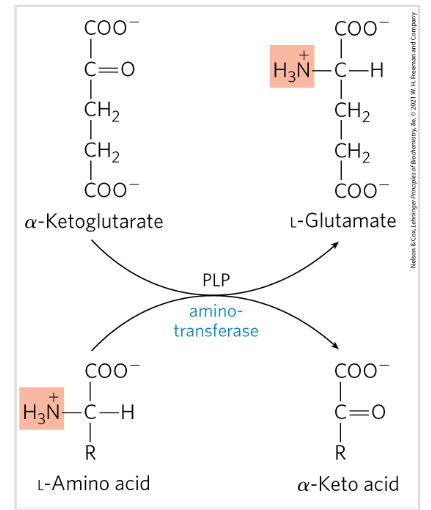
Degradation of aa in the liver- transamination
Removal of the alfa amino group from an aa to alfa keto glutarate→ transaminases/aminotransferases
Transamination- alfa amino group is transferred to the alfa carbon atom of alfa keto glu→ amino group acceptor
Requires PLP=pyridoxal phosphate → coenzyme of pyridoxine
Glutamate =FINAL product=nitrogen carrier
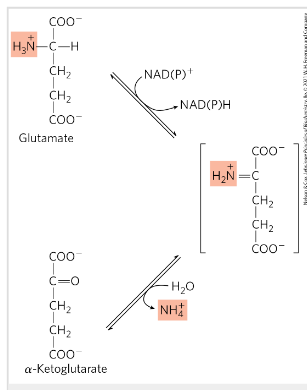
Degradation of aa in the liver- oxidative deamination
Removal of the amino group of glutamate releasing free ammonia NH3
Glutamate from transamination in cytosol is transferred to mitochondria in liver (need to be converted to free ammonia)
Cofactor NAD+/NADP+
Reaction catalyzed by L-glutamate dehydrogenase
Releases ammonia from glutamate and forms alfa ketoglutarate
Combined action of aminotransferase and glutamate dehydogenase is called
Transdeamination
Alfa keto glutarate is formed and can be used in citric acid cycle
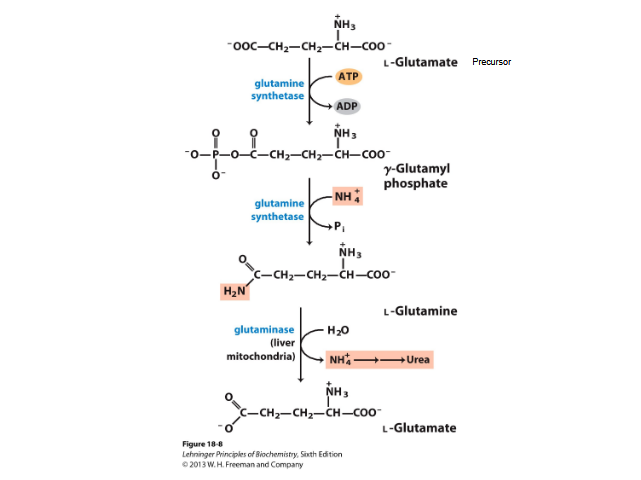
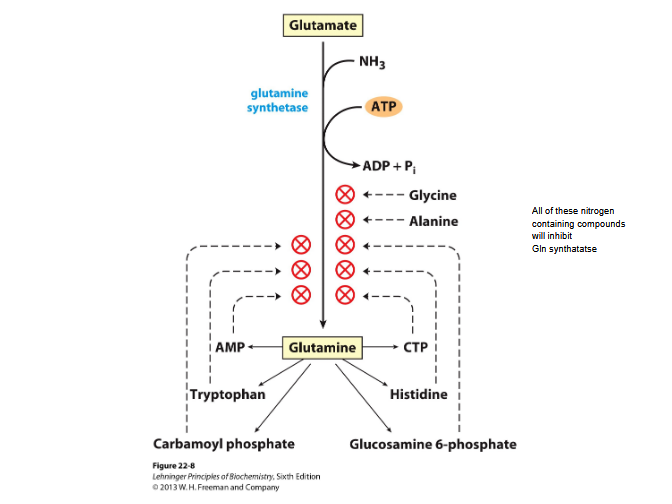
Glutamine transports ammonia
NH3 + glutamate → glutamine by the action of glutamine synthase
2 step process requiring ATP
Detoxifies ammonia
Excess glutamine→ breaks down to glutamate and ammonium NH4+→ by glutaminase
Then ammonia → urea in liver
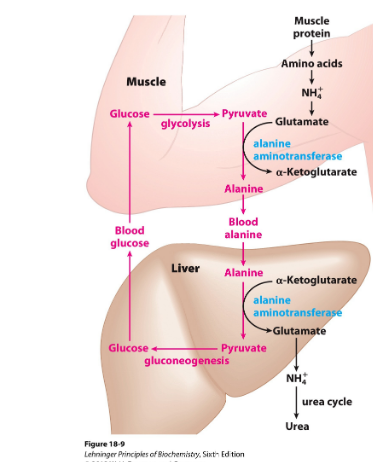
Alanine transports
Muscles produce pyruvate, lactate and ammonia during anaerobic contracting muscles→ must go to liver
Lactate + pyruvate→ incoorperated into glucose→ muscles
Ammonia is converted to urea for excretion
amino groups from muscle to liver
Gluconeogenesis in liver
Picture: glucose-alanine cycle
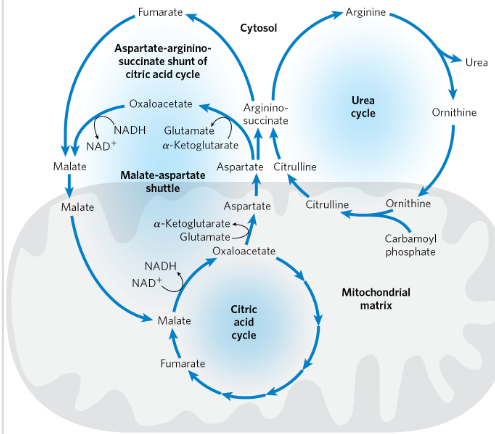
Urea cycle- excretion of N
occur in both mitohondria and cytosol
Urea is produced from ammonia in 5 steps
In liver
Uses 2 nitrogen sources: carbomoyl phosphate + aspartate
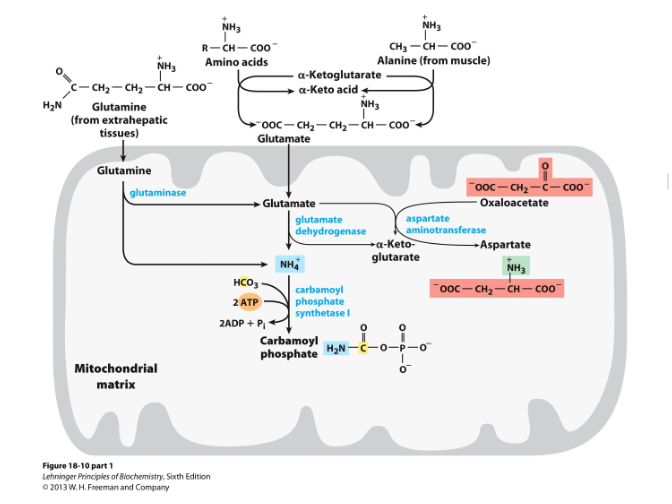
4 steps of urea cycle:
Ornithine + carbamoyl phosphate → citrulline (in mitochondria)
Citrulline → argininosuccinate (in cytosol; second nitrogen added from aspartate)
Argininosuccinate → arginine + fumarate
Arginine → urea + ornithine (ornithine re-enters mitochondria to continue the cycle)
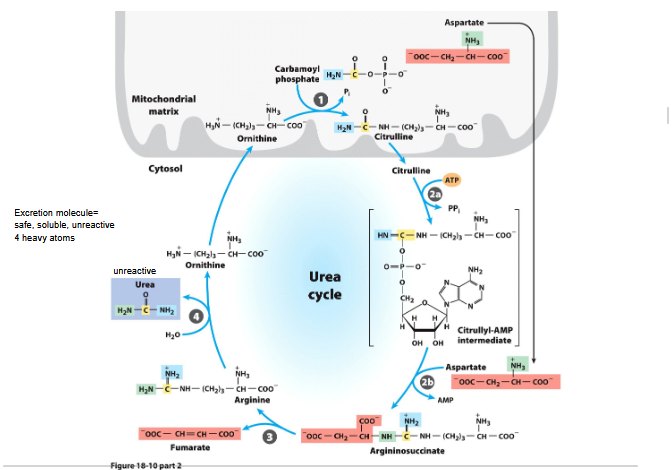
Citric acid cycle and ureas cycle is linked
Fumarate produced in argininosuccinase reaction→ intermediate for the citric acid cycle
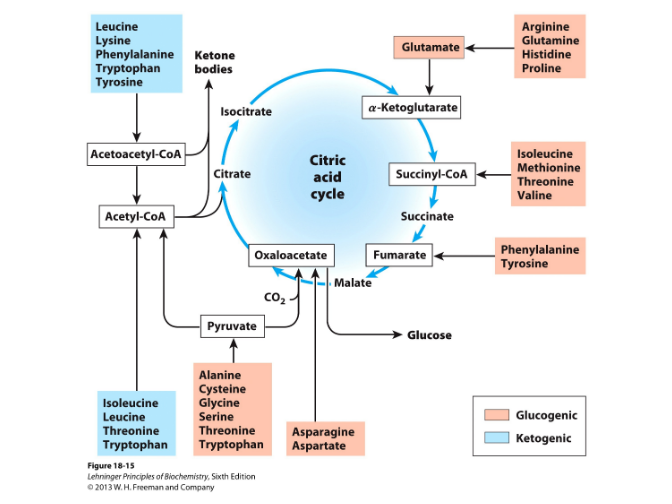
Intermediates that can enter citric acid cycle
degradation of aa carbon skeletons (alfa-keto acids)- to intermediates of the citric acid cycle→ gluconeogenesis + ketogenesis
6 major intermediates: pyruvate, acetyl CoA, alfa ketoglutarate, succinyl CoA, fumarate, OAA
ketogenic=aa degraded to form ketone bodies
glucogenic aa=degraded to form pyruvate, alfaketoglutarate, succinyl CoA, fumarate, OAA→ glucose and glycogen
Degradation of nucleotides into
Degradation of nucleotides
purines→ uric acid
pyrimidines→ succinyl COA
Synthesis of aa and nucleotides
By glutamine synthase
Regulated allosteric and by adenylyation- nitrogen compounds can inhibit Gln synthase
Reaction: Glu + ATP + NH4+→ Gln + ADP + Pi + H+
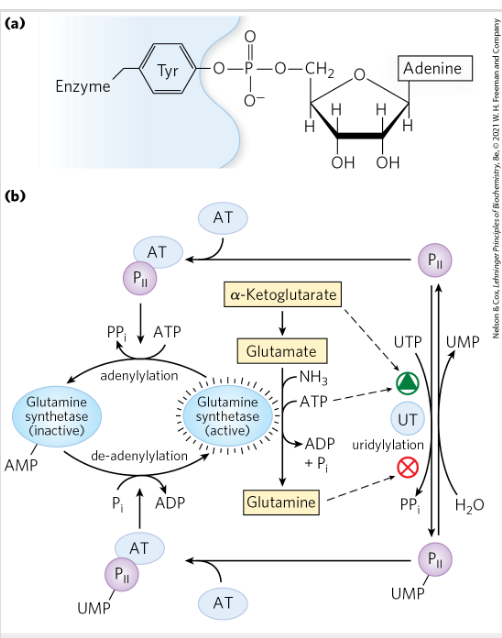
Protein P2- activates adenylyation of glutamine synthase
It activates AT that adds an adenylyl group (–AMP) to glutamine synthetase. This process is called adenylylation.
This modification regulates the activity of glutamine synthetase, which controls how cells use ammonia to synthesize amino acids
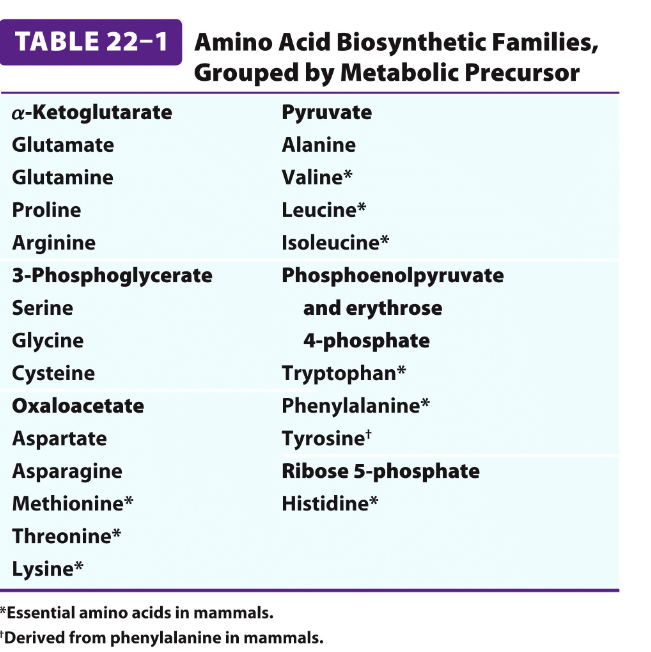
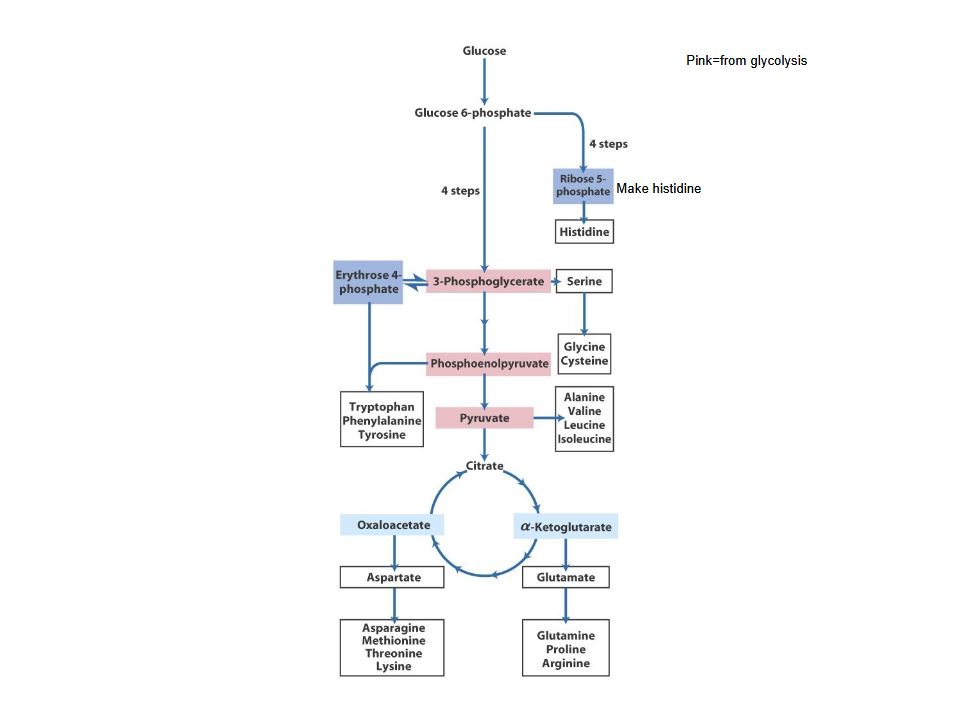
AA derived form intermediates in three pathways
All 20 standard amino acids in humans are synthesized from key intermediates of central metabolic pathways
Citric acid cycle- provides intermediates like alfaketoglutarate, OAA, fumarate
Glycolysis→ supplies intermediates such as 3-phosphoglycerate, pyruvate, phosphoenolpyryvate
Pentose phosphate pathway→ produces ribose-5-phosphate and more
Intermediates of the three pathways
*) aKG
pyruvate
3-phoshpoglycerate
PEP
Erythrose 4 -phosphate (PPP)
OAA
Ribose phosphate (PPP)
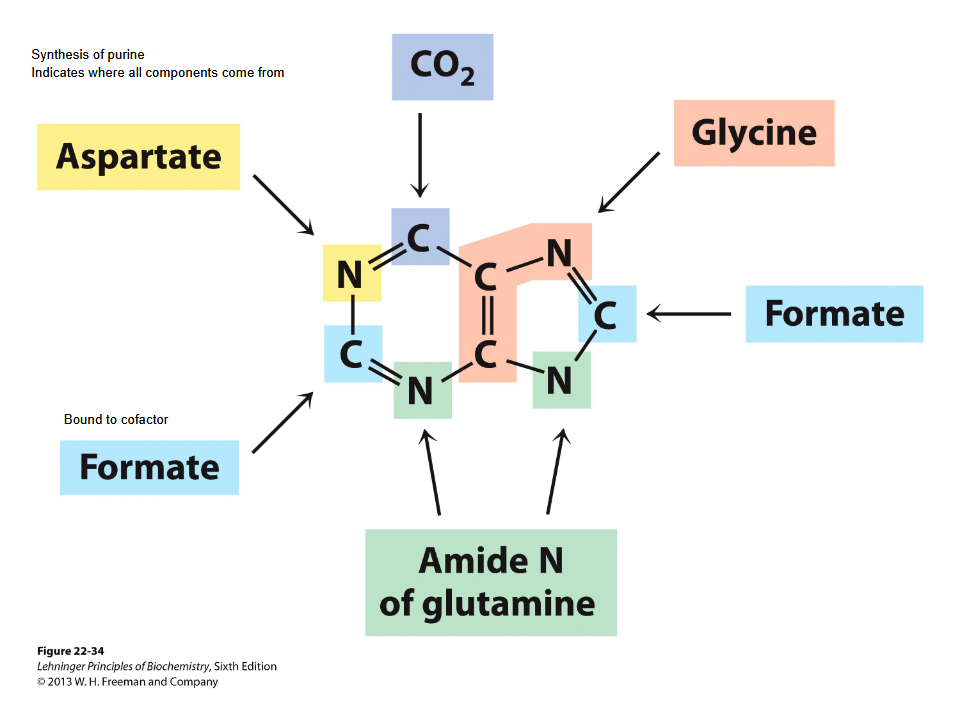
SYnthesis of nucleotides- 2 pathways
) salvage pathway -
using available riboses
and bases-recycles free bases or nucleosides derived from the breakdown of DNA and RNA.
*) de novo pathway->
synthesized from ribo 5
phosphate + CO2-R + NH3
From scratch-using small precursor molecules such as amino acids, carbon dioxide, and ribose-5-phosphate.