Exam 2 Microbio
1/130
Earn XP
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
131 Terms
“normal“ growth conditions
sea level
20-40 C
0.9% salt
ample nutrients
Environmental Limits on Microbial Growth
temperature
pH
osmolarity
oxygen
pressure
Hyperthermophiles
Organisms that thrive in extremely high temperatures, above 80 degrees Celsius. They are found in geothermal areas like hot springs and hydrothermal vents.
Thermophiles
grow between 50-80 degrees celsius
Mesophile
grow between 15-45 C
Psychrophile
grow below 15 degrees C
Alkaliphile
growth above pH 9 (alkaline)
Neutralophile
grow between 5-8 pH
Acidophile
grow below 3 pH
Halophile
growth in high salt (>2M NaCl)
Aerobe
growth only in O2
Faculative
growth with or without O2
Microaerophile
growth only in small amounts of O2
Anaerobe
growth only without O2
Barophile
growth at high pressure (above 380 atm)
Barotolerant
growth between 10 and 495 atm (high pressure but not extremely high)
hypotonic solution effect
cause cell to swell and burst
hyperyonic solution effect
cause cell to shrivel (plasmolysis)
Microbes that grow at temperatures between 40°C and 80°C are called:
A. Psychrophiles
B. Mesophiles
C. Thermophiles
D. Extreme Thermophiles
Thermophiles
Bacteria cannot grow in solutions with very high concentrations of sugar because:
A. Bacteria cannot digest pure sugar
B. Sugar raises the solution’s osmolarity
C. Sugar alters the solution’s pH
Sugar raises the solution’s osmolarity
Weak acids can pass through membranes which:
Disrupts cell pH homeostasis, and kills cells. This makes them good food preservatives.
How do anaerobes survive?
Must use a terminal electron acceptor other than O2
Three oxygen-removing techniques are used today
Special reducing agents
(thioglycolate) or enzyme
systems (Oxyrase) can be
added to ordinary liquid
media
An anaerobe jar
- O2 is removed by a reaction
catalyzed by palladium
- Newer methods ‘Gaspak’,
don’t need palladium
catalyst
An anaerobic chamber
with glove ports
- O2 is removed by vacuum
and replaced with N2 and
CO2
Eutrophication
the sudden infusion of large quantities of a formerly limiting nutrient, which can lead to a bloom of microbes
sterilization
killing of all living organisms
disinfectant
killing or removal of pathogens from inanimate objects
antisepsis
killing or removal of pathogens from a surface of living tissue
sanitation
reducing the microbial population to safe levels
Pasteurization vs Flash Pasteurization
63°C for 30 minutes vs. 72°C for 15 seconds
Ultra High Temperature
150°C for 3 seconds, sterilizes (all bacteria killed)
Used for creamers
Autoclave
steaming (heat and moisture) under pressure (kills bacteria and endospores)
121°C, 15 psi (2 atm) for 20 min
A number of factors influence the efficacy of a given chemical agent, including:
- The presence of organic matter
- The kinds of organisms present
- Corrosiveness
- Stability, odor, and surface tension
Probiotics
“good bacteria” that displace disease causing organisms from tissues
Bacteriophage
Viruses that attack bacteria (do not harm eukaryotes)
which microbe is most resistant to killing by environmental stress agents?
prions
To sterilize heat-labile (sensitive) solutions, one should use
A. dry heat.
B. an autoclave.
C. membrane filtration.
D. Pasteurization.
Membrane filtration
Ethanol can be used as an effective bactericide when used at a concentration of
A. 70%.
B. 50%.
C. 40%.
D. 100%
70%
Generally, the simpler the organism
the smaller the genome
Purine
A and G
Pyrimidine
C, T and U
Nucleotides are connected by
a 5’-3’ phosphodiester bonds
why is GC pair is stronger than AT pair?
GC has 3 hydrogen bonds instead of the two that AT has
If the sequence of one strand of DNA is
5’ TCGATC 3’, what is the sequence of the complementary strand?
a) 5’ CTAGCT 3’
b) 5’ GCTAGC 3’
c) 5’ AGCTAG 3’
d) 5’ GATCGA 3’
C
what regulates supercoiling?
topoisomerases
Plasmids
extrachromosome DNA molecules.
- Much smaller than the chromosome
- Usually circular
- Need host proteins to replicate
- Low-copy-number plasmids
- High-copy-number plasmids
Plasmid replication
rolling circle (unidirectional) replication, complementary strand synthesized as unroling occurs
Used by many bacteriophage viruses and mammalian DNA viruses
Analysis of the genome of a newly discovered bacterial strain reveals that it is composed of a double-stranded DNA molecule containing 16% thymine. Based on this information, what would you predict the percentage of cytosine to be?
A. 34%
B. 68%
C. 16%
D. 32%
34%
When the DNA replicates, how is the newly made strand related to its template strand?
A. The two strands have identical sequences and are antiparallel to each other
B. The two strands have complementary sequences and are parallel to each other
C. The two strands have complementary sequences and are antiparallel to each other.
D. The two strands have identical sequences and are parallel to each other.
E. The two strands have identical sequences and are antiparallel to each other, except that U replaces T.
C. The two strands have complementary sequences and are antiparallel to each other.
Replication, transcription, and translation all have 3 major steps
initiation
elongation
termination
RNA Polymerase
Enzyme responsible for transcription in cells. It synthesizes RNA molecules using a DNA template. Helps in the production of mRNA, tRNA, and rRNA.
Core polymerase: a2, b, b’, w
sigma subunit guides RNA Polymerase to target DNA sequence
Transcription initiation steps
sigma (as part of RNA holoenzyme) scans and binds starting sequence
Binding to the promoter sequence allows for the closed complex to form
RNA polymerase unwinds DNA and begins transription
sigma factor leaves the complex
The unwinding of DNA ahead of the moving complex forms a
17 base pair transcription bubble
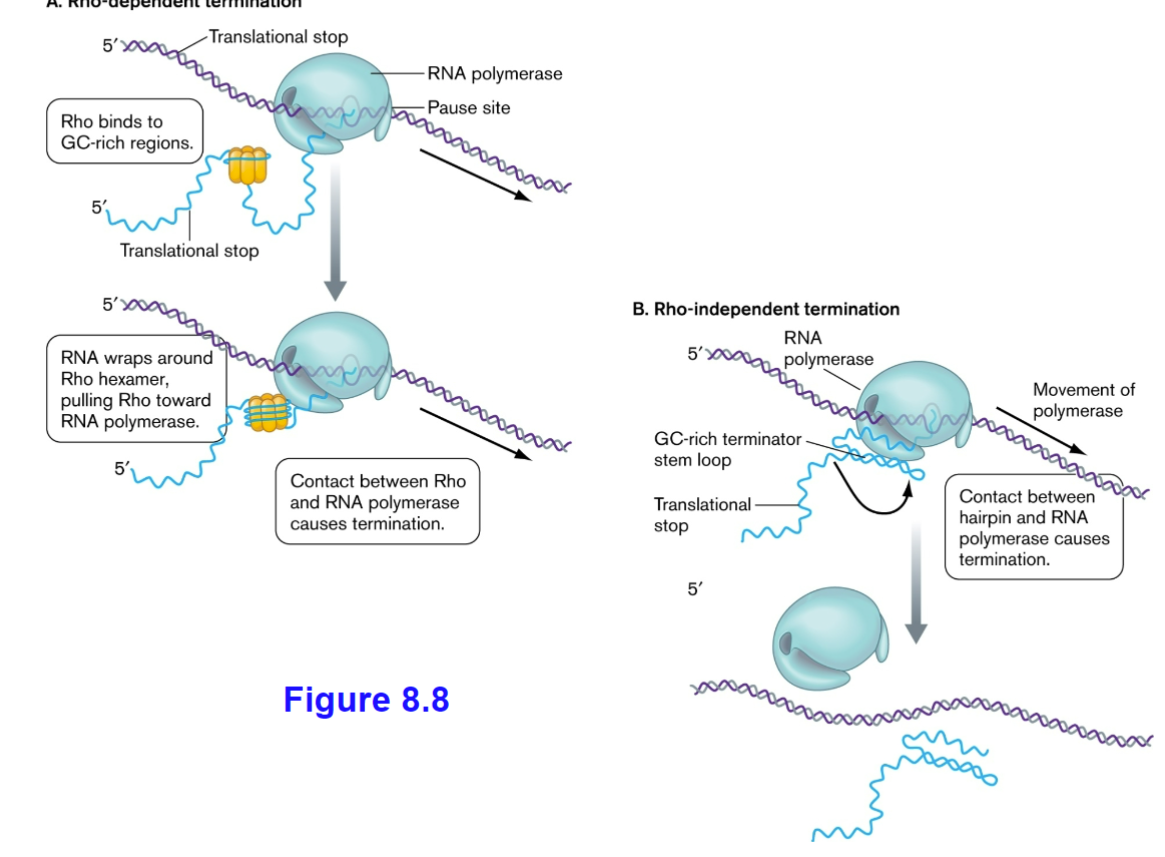
Rho-dependent vs rho-independent termination
Termination of transcription in bacteria can be rho-dependent or rho-independent. Rho-dependent termination relies on a protein called Rho, which binds to the mRNA and causes the RNA polymerase to detach from the DNA template.
Rho-independent termination, on the other hand, occurs when a specific DNA sequence forms a hairpin loop followed by a string of uracil bases in the mRNA. This structure causes the RNA polymerase to pause and eventually dissociate from the DNA.
Antibiotics that Affect Transcription
Rifamycin B
Actinomycin D
What processes are common to replication, transcription, and translation?
A. Initiation, base-pairing, termination
B. Initiation, elongation, termination
C. Elongation, use of GTP for energy, use of accessory proteins
B
The sigma factor is directly required for
a) Transcription initiation
b) Transcription elongation
c) Translation initiation
d) Translation elongation
Transcription initiation
Number of possible codon sets
64 total
3 stop
61 amino acid coding sets (only 20 amino acids)
A tRNA molecule has two functional regions:
Anticodon: hydrogen bonds with the mRNA codon specifying an amino acid
3′ (acceptor) end: binds the amino acid
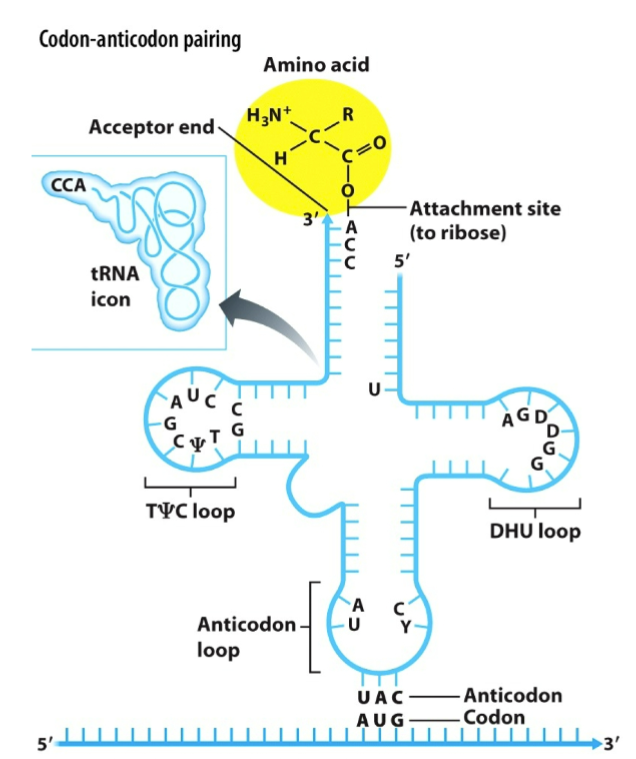
The charging of tRNAs is carried out by a set of enzymes called
aminoacyl-tRNA synthetases
In prokaryotes, the subunits are
30S and 50S and combine to form the 70S ribosome
3 binding sites of 70s ribosomes:
1. A (acceptor) site: binds incoming aminoacyl-tRNA
2. P (peptidyl-tRNA) site: harbors the tRNA with the growing polypeptide chain
3. E (exit) site: binds a tRNA recently stripped of its polypeptide
Antibiotics That Affect Translation
Streptomycin: inhibits 70S ribosome formation
Tetracycline: inhibits aminoacyl-tRNA binding to the A site
Chloramphenicol: inhibits peptidyltransferase
Puromycin: triggers peptidyltransferase prematurely
Erythromycin: causes abortive translocation
Fusidic acid: prevents translocation
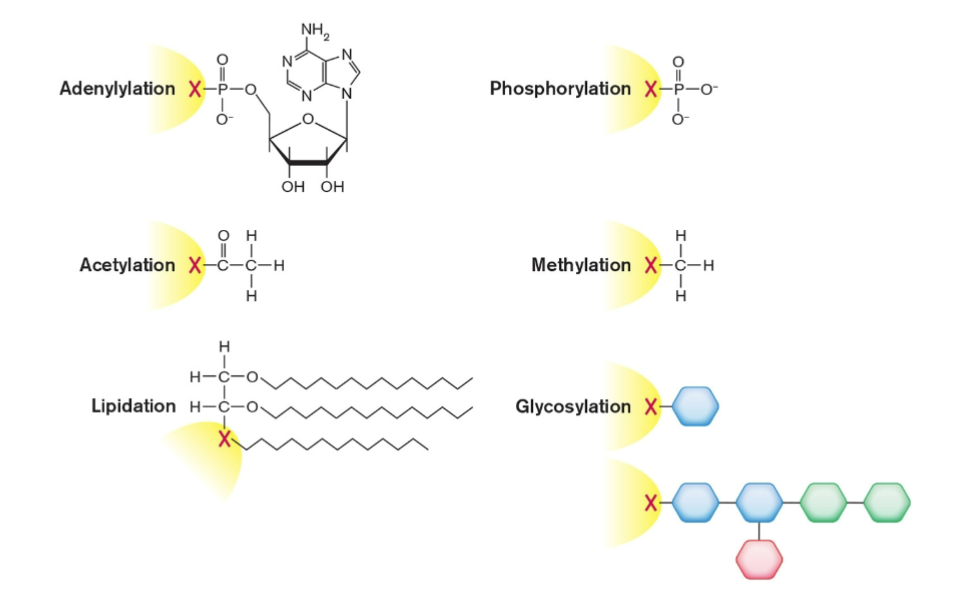
Protein Modification
Enzymes modify translated protein
fMet removed from N-terminus
Small groups added to amino acids
Phosphoryl groups added
Methyl groups added
Adenylate groups added
Protein may be cleaved
Protein may be refolded by helping enzymes
signal sequences
target proteins for transportation
found on N terminus end of proteins
The types of RNA that physically catalyzes the formation of polypeptides is:
a. sRNA
b. tRNA
c. rRNA
d. mRNA
rRNA
Which of the following RNAs is used as templates for protein translation?
a) mRNA
b) tRNA
c) rRNA
d) tmRNA
mRNA
Chaperones are proteins that
a) Export other proteins out of the cell
b) Help other proteins fold properly
c) Degrade misfolded proteins
d) Bind other proteins to inactivate them
Help other proteins fold properly
Signal sequences are found
a) At the 3′-end of an mRNA molecule
b) At the 5′-end of an mRNA molecule
c) In the 50S ribosomal subunit
d) At the C-terminus of a protein
e) At the N-terminus of a protein
at the N-terminus of a protein
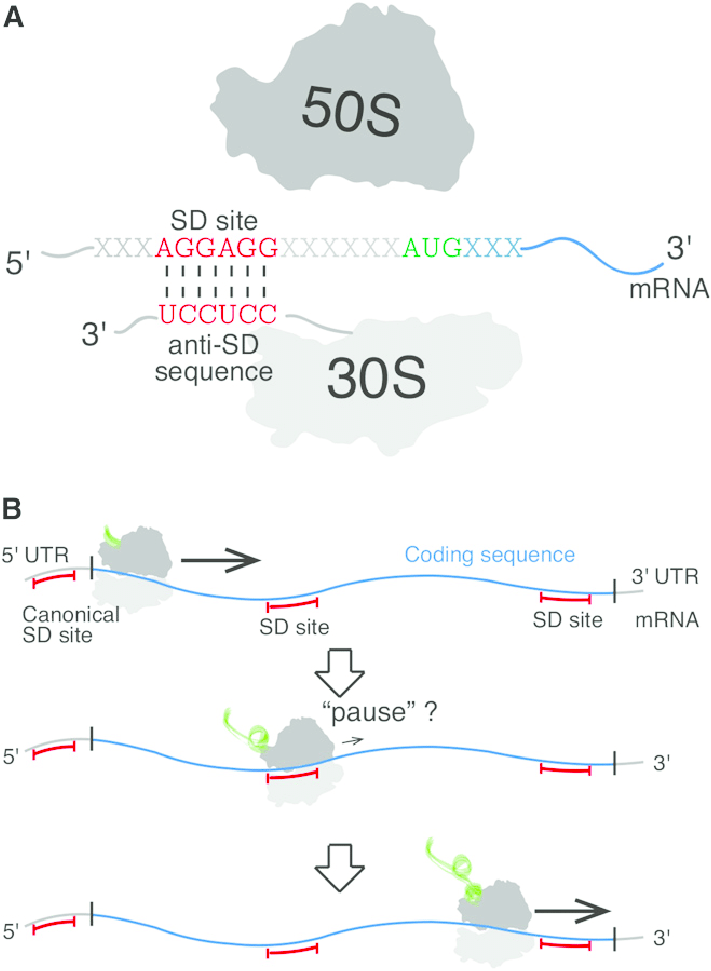
Shine-Dalgarno sequences
is complementary to a sequence at the 3′ end of 16S rRNA of the 30S subunit, it directs the start of translation
mutation
a heritable change in DNA
Point mutations
change of a single base (not deleted or inserted)
transition
purine→purine or pyrimidine→pyrimidine
transversion
purine↔pyrimidine
insertion or deletion
addition/subtraction of one or more bases
inversion
DNA is flipped in orientation
reversion
DNA mutates back to original sequence
silent mutation
mutation does not change the amino acid
missense mutation
mutation changes the coded amino acid
nonsense mutation
mutation changes to a stop codon
Frame-shift mutation
mutation changes the ORF of a gene
Mutations Arise in Diverse Ways
1. Tautomeric shifts in DNA bases that alter base-pairing properties
2. Oxidative deamination of bases
3. Formation of apurinic sites
4. Damage caused by reactive oxygen species
Ames Test
DNA repair
Error-proof repair
Error-prone repair
Error-proof repair
pathways, which prevent mutations
Methyl mismatch repair
photoreactivation
nucleotide excision repair
base excision repair
recombinational repair
Error-prone repair
pathways, which risk introducing mutations
Operate only when damage is so severe that the cell has no other choice but to die
Methyl mismatch repair
based on recognition of the methylation pattern in DNA bases.
Uses methylation of the parental strand to discriminate from newly replicated DNA
Photoreactivation
The enzyme photolyase binds to the pyrimidine dimer and cleaves the cyclobutane ring
Nucleotide excision repair
An endonuclease removes a patch of single-stranded DNA containing certain types of damaged bases, including dimers.
Base excision repair
Specialized enzymes can recognize specific damaged bases and remove them without breaking the phosphodiester bonds.
This AP site allows DNA pol I to synthesize a replacement strand containing the proper base
Recombinational repair
A single-stranded segment of the undamaged daughter strand can be used to replace a gap in the damaged daughter strand
SOS (“Save Our Ship”) repair
Induced by extensive DNA damage
- RecA coprotease activity stimulates autodigestion of the LexA repressor.
- Expression of many DNA repair enzymes
- Among them, 2 “sloppy” DNA polymerases that lack proofreading activity
- However, the cell has no other option but to “mutate or die.”
Taking up foreign DNA can be beneficial.
• Imported DNA can be used as an alternative food source.
• Repair damaged chromosomes
• Drive genome evolution
Why do species undergo natural transformation?
Use indiscriminate DNA as food
Use specific DNA to repair damaged genomes
Acquire new genes through horizontal gene transfer
Conjugation
“bacterial sex”, transfers DNA through sex pilus
F’ factor
F plasmid contains extra genes (in addition to genes for pilus and transfer)
Transfers extra genes to recipient
Hfr conjugation
F factor integrates on bacterial chromosome, tries to transfer entire chromosome (Requires 100 minutes for E. coli). Transfers genes in order and can be used to determine order of genes on chromosome
transduction
the process in which bacteriophages carry host DNA from one cell to another
Which of the following sequences is not a restriction enzyme cut site?
A) GGATCC
B) GAATTC
C) CTTAAG
D) GGGCCC
E) CCCTTT
E) CCCTTT
CRISPR
consist of repeats and spacers that do not encode proteins, but near them lie CRISPR-associated gene families that do encode proteins
in an Ames Test, a very strong mutagen will cause the appearance of:
a. Many colonies over the plate, with a clear space close to the disk of mutagen.
b. A few colonies throughout the plate up to the edge of the disk.
c. Many colonies covering the whole plate, extending up to the disk.
a. Many colonies over the plate, with a clear space close to the disk of mutagen.