Chapter 3, Protein Structure and Function: Hierarchical Structure of Proteins, Protein Fold
1/86
Earn XP
Description and Tags
Bolded vocab words in the slides defined by the glossary
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
87 Terms
Primary Structure
In proteins, the linear arrangement (sequence) of amino acids within a polypeptide chain.
Secondary Structure
In proteins, local folding of a polypeptide chain into regular structures including the alpha helix, Beta sheet, and Beta turns
Local 3D shapes held together by H bonding
Tertiary Structure
In proteins, overall three-dimensional form of a polypeptide chain, which is stabilized by multiple noncovalent interactions between side chains.
Full 3D shape of a single peptide held together by several types of interactions.
Quaternary Structure
The number and relative positions of the polypeptide chains in multimeric (multisubunit) proteins.
Association of two + folded peptides (subunits) as a complex performing a single function.
Supramolecular complexes
Large multi-subunit complexes that perform several interrelated functions.
Structure
Organizing the genome, organelles, cytoplasm, protein complexes, and membranes in 3D space
Regulation
Controlling protein activity and gene expression
Signaling
Monitoring the environment and transmitting information
Transport
Moving small molecules and ions across membranes
Enzyme Activity
Catalyzing chemical reactions
Motors
Generating force for movement
Translation reads from
5’ → 3”
N-terminus to C-terminus
Native conformation
normal, functional conformation
Non-native conformations
Misfolded proteins have aberrant functions or aggregate causing defects in cell function or cell death.
Amyloid fiber is a pole of aberrant proteins.
Dalton
Unit of molecular mass approximately equal to the mass of a hydrogen atom (1.66×10^-24g).
Alpha a helix
Common protein secondary structure in which the linear sequence of amino acids is folded into a right-handed spiral stabilized by hydrogen bonds between carboxyl and amide groups in the backbone.
Right handed helix ~3.6 aa per turn.
each residue is hydrogen bonded to the amino acid 4 residues above and below it.
R group is on the outside. Prolines can’t participate in hydrogen bonding and are very uncommon in alpha helices, they disrupt it sterically.
Steric
Relating to the spatial arrangement of atoms in a molecule, especially as it affects chemical reactions.
Beta B sheet
A flat secondary structure in proteins that is created by hydrogen bonding between the backbone atoms in two different polypeptide chains or segments of a single folded chain.
Large side chains can sterically interfere with sheet formation
Strands can run parallel or antiparallel
R groups alternatively project above and below the plane.
Beta B turn
A short U-shaped secondary structure in proteins.
Change the direction of the peptide backbone by 180˚, allowing the peptide chain to fold back onto itself.
composed of 4 residues.
alpha carbons of the 1st and 4th residues are hydrogen bonded.
Usually contain glycine and proline.
Structural Motif
A particular combination of two or more secondary structures that form a distinct three-dimensional structure that appears in multiple proteins and that often, but not always, is associated with a specific function.
Nucleation
The initial process that occurs in the formation of a move complex and stable arrangement.
Coiled Coil motif
Two or more alpha helices wound tightly around each other, stabilized by hydrophobic interactions.
Each alpha helix contains a 4 amino acid repeat called a heptad repeat.
The 1st and 4th redidues of each repeat are aliphatic (hydrophobic but not aromatic) usually leucine and valine.
Stabilized by hydrophobic interactions.
EF hand
A type of helix-loop-helix structural motif that occurs in many Ca²+ binding proteins such as calmodulin.
Two alpha helices held at right angles to one another, joined by a loop that forms the angle.
Proteins with EF hands are sensitive to change in Ca2+ levels
Conformation/ function is controlled by regulated release of Ca2+ from the ER.
Helix-Loop-Helix
A conserved DNA-binding structural motif, consisting of two alpha helices connected by a short loop, that is found in many dimeric eukaryotic transcription factors.
Basic Helix-loop-helix
Important helix loop helix motif. Proteins with bHLHs usually bind DNA and include transcription factors.
Alpha helices connected by flexible linker. Two helices two separate functions.
Transcription Factor
General term for any protein, other than RNA polymerase, required to initiate or regulate transcription in eukaryotic cells. General factors, required for transcription of all genes, participate in formation of the transcription-preinitiation complex near the start site. Specific factors stimulate (activators) or inhibit (repressors) transcription of particular genes by binding to their regulatory sequences.
Protine Dimerization
Is a biological process where two proteins interact to form a functional complex.
Zinc-finger
Motif with one or more Zn2+ ions coordinated an alpha helix and a small beta sheet to stabilize the fold.
Function as interaction regions with other proteins, DNA, RNA, or lipids.
Protein Domain
Distinct regions of a protein’s three-dimensional structure. A functional domain exhibits a particular activity characteristic of the protein; a structural domain is ~40 or more amino acids in length, arranged in a distinct secondary or tertiary structure; a topological domain has a distinctive spatial relationship to the rest of the protein.
Secondary structures and structural motifs that fold and pack together forming compact, local, semi-independent 3D units.
Domain
A region of protein that has a distinct, and often independent, function or a structure, or that has a distinct topology relative to the rest of the protein.
Functional Domains
Exhibit specific enzymatic activity, usually independent of other regions of the protein, even when isolated from the rest of the protein.
Structural Domain
Region of 40+ residues arranged as a single, stable, distinct structure often comprised of one or more secondary structures.
Topological Domain
Regions defined by their spatial relationship to the rest of the protein
e.g., membrane spanning proteins have extracellular, membrane embedded and cytoplasmic domains.
Homolog
A protein that shares a common ancestor, and therefore is similar in sequence and/or structure, with another protein.
Families
Groups of proteins with common ancestry (homologs) and families are groups within super families.
AlphaFold
AI software created by Google’s Alphabet can predict protein structure and folding.
Globular Proteins
Generally water-soluable, compactly folded structure.
Fibrous proteins
large, elongated, often stiff molecules.
Integral Membrane Protein
Any protein that contains one or more hydrophobic segments embedded within the core of the phospholipids bilayer; also called transmembrane protein.
Intrinsically disordered region (IDR)
A segment (region) of a protein that is very flexible, with no fixed, well-ordered conformation. If the entire protein has no fixed conformation, it is called an intrinsically disordered protein (IDP).
No well-ordered structure in their native conformation.
Rich in polar amino acids and proline.
Interact with multiple partner proteins.
Induced fit
The interaction between an intrinsically disordered protein and a specific well-ordered partner results in conformational change that allow the molecules to interact with greater affinity for one another.
Chaperone
Collective term for two types of proteins - molecular chaperones and chaperonins - that prevent misfolding of a target protein or actively facilitate proper folding of an incompletely folded target protein, respectively.
Molecular chaperones
Bind short segments of a nascent protein and stabilize unfolded or partly folded regions, preventing aggregation or degradation.
ATP-dependent proteins
Chaperonins
Form folding chambers into which all of a nascent protein can be sequenced without interference from other molecules.
Supramolecular complexes
Perform several functions al related to some very complex process, like RNA transcription.
Nascent protein
A protein that is being translated or has not yet adopted its fully folded native conformation. (Nascent: something just coming into existence and beginning to display signs of future potential.)
Peptidyl-proline isomerases
Enzymes that catalyze cis to trans proline isomerization in mature proteins.
Heat-Shock Proteins (HSPs)
Proteins that perform several processes that protect cells from the consequences of unfolding a large fraction of cellular proteins.
Ligand
Small molecule to which a protein binds
often results in a protein conformation change
Ribozyme
An RNA molecule with catalytic activity. Ribozymes function in RNA splicing and protein synthesis.
Substrate
Molecule that undergoes a charge in a reaction catalyzed by an enzyme.
Cofactors
Inorganic atoms or molecules, mostly metal ions, which participate directly in catalysis through ionic interactions with the substrate- the molecules bing acted upon.
Coenzymes
Organic enzyme conjugated, usually derived from vitamins, that generally function as intermediate carriers of electrons, specific atoms or functional groups that are transferred in the catalyzed reaction.
Active Site
Specific region of an enzyme that binds a substrate molecule(s) and promotes a chemical change in the bound substrate.
Two important components substrate binding site, and catalytic site.
Substrate Orientation
Multiple substrates are brought together in the correct orientation to catalyze the reaction.
Changed Substrate Reactivity
The substrate may be influenced by amino acid side chains in the active site that alter their chemical properties.
Induced Strain on the Substrate
Enzyme induced conformation change of the substrate.
Enzyme Kinetics
The study of the rates of enzymes catalyzed reactions under variable experimental conditions.
Saturated
Referring to a compound (e.g., fatty acid) in which all the carbon-carbon bonds are single bonds.
Vmax. Maximal Velocity
Parameter that describes the maximal velocity of an enzyme-catalyzed reaction or other process such as protein-mediated transport of molecules across a membrane.
Kcat
Catalytic constant (Turnover number) the number of reaction scatalyzed by a single enzyme per second when operating at saturation. Kcat= Vmax/ [E]total
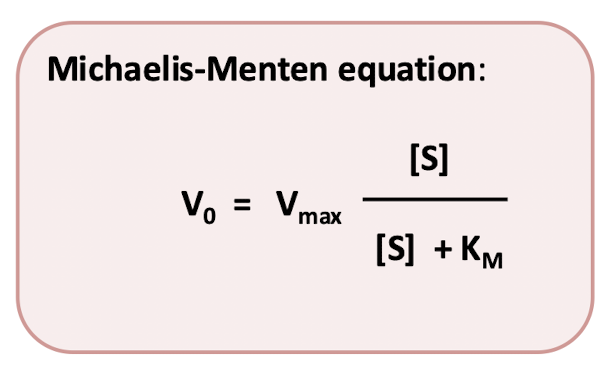
Michaelis Constant (Km)
A parameter that describes the affinity of an enzyme for its substrate and equals the substrate concentration that yields the half-maximal reaction rate. A similar parameter describes the affinity of a transport protein for the transported molecule or the affinity of a receptor for its ligand.
equal to substrate concentratino at ½ Vmax.
If Km is large low affinity for substrate.
If Km is small high affinity for substrate.
Serine Proteases
A large family of enzymes that catalyze the cleavage of specific peptide bonds via hydrolysis reactions.
Trypsin
preferentially cleaves the peptide bond C-terminal to residues with positively charged side changes (arginine and lysine).
Main digestive enzyme produced in the small intestine.
Catalytic Triad
Three precisely positioned residues cooperate to attach the peptide bond of the substrate.
Our ex Aspartic acid - Histidine- Serine all hydrogen bond to each other
Asp bonded to His makes His a stronger base and it steals Ser’s protons which enhances Ser’s nucleophile capacity.
Regulation of Proteins
Cells can increase/ decrease the steady state levels of a protein by altering its rate of synthesis/ degradation
Change the intrinsic activity of the protein
change the location or concentration within subcellular regions of the protein.
Protein Degredation
Regulates life span of intracellular proteins, removes damaged proteins that may be toxic to the cells.
Proteasome
Large multifunctional protease complex in the cytosol that degrades intracellular proteins marked for destruction by attachment of multiple ubiquitin molecules.
Control ow long specific proteins are active in a cell or degrade misfolded proteins important in ER system.
Ubiquinone (coenzyme Q (CoQ))
A hydrophobic small, organic molecule that carries two protons and two electrons in electron-transport pathways, cycling between the oxidized (ubiquinone) and reduced (dihydroquinone) forms.
Ubiquitinylation
The enzymatic, covalent attachment of the small protein ubiquitin to a protein.
Ubiquitin
A small protein that can be covalently linked to other intracellular proteins, thereby tagging these proteins for degradation by the proteasome, sorting to the lysosome, or alteration in the function of the target protein.
Signal to proteasomes to degrade a protein.
Allosteric regulation
A regulatory ligand binds a protein in a specific allosteric binding site which induces a conformational change in the protein that may activate or inhibit the activity of the protein.
Feedback
A regulatory process in a signaling pathway where one of the later components of the pathway, when activated, inhibits (negative feedback) or activates (positive feedback) an earlier step in the pathway.
Feedback Repression
An essential feature of most signal transduction pathways, in which an end product of a signaling pathway blocks an early step in that pathway.
Feedforward Activation
An early product may bind and allosterically activate an enzyme at a later step in the pathway.
Cooperativity
Unique allosteric regulation seen in protein complexes with multiple binding sites for the same ligand. Can be positive or negative.
Irreversible inhibitors
bind tightly to the enzyme
Competitive indibitors
Structural analogs of the enzyme substrate. Bind enzyme active site and inhibit normal substrate from being bound.
Non-competitive inhibitors
Allosteric inhibitors. Bind at a location other than the activation site, altering enzyme conformation and ability to bind substrate or catalyze reaction.
Switch protein
Activates other proteins.
GTPase superfamily
Group of intracellular switch proteins that cycle between an inactive state with bound GDP and an active state with bound GTP. Includes the Galpha subunit of trimeric (large) G proteins monomeric (small) G proteins (e.g., Ras, Rab, Ran, and Rac), and certain elongation factors used in protein synthesis.
G protein- coupled receptor (GPCR)
Member of a large class of cell-surface signaling receptors, including those for epinephrine, glucagon, and yeast mating factors. All GPCRs contain seven transmembrane alpha helices. Ligand binding leads to activation of a coupled trimeric G protein, thereby initiating intracelular signaling pathways.
Kinase
An enzyme that transfers the terminal (gama) phosphate group from ATP to a substrate. Protein kinases, which phosphorylate specific serine, threonine, or tyrosine residues, play a critical role in regulating the activity of many cellular proteins.
Protein Kinase (PK)
An enzyme that chemically adds a phosphate group, usually the gamma phosphate from ATP, to a serine, threonine, or tyrosine residue of another protein.
Protein Phosphatase
An enzyme that chemically removes a phosphate group from another protein by hydrolysis.
Prohormones
must be cleaved into active hormones before release from the cell
Symogens
inactive enzyme precursors that have not yet been activated by proteolytic cleavage.