Unit 4: eukaryotic gene control
1/35
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
36 Terms
Eukaryotic cells tend to have gene regulation in ____ potential points
many:
transcription
post transcription but before translation
translation
post translation
example of eukaryotic gene control during transcription
transcription factors
enhancers
histones
etc
example of eukaryotic gene control post transcription but before translation
significant process of turning mRNA into mature mRNA
certain areas (introns) of the mRNA are cut out
this is a type of regulation bc if they are cut out they will not be expressed (won’t be translated)
Example of eukaryotic gene control during translation
Eukaryotic Initiation Factor 2 (eIF-2) helps translation get started
eIF-2 can be regulated
if eIF-2 is phosphorylated (added phosphate group) its shape changes and can no longer initiate transcription
no protein is made = no gene expression
Example of eukaryotic gene control post translation
chemical groups can be added or removed from proteins
this can change where the proteins end up located or how they function
therefore impacting their expression
eg. environmental factors can influence this (UV, low nutrients, etc)
Ubiquitin
post translation gene control
regulatory protein
ubiquitin tagging = protein degredation (cell death)
what does altering the rate of transcription do?
Its a common method of regulating the expression of eukaryotic genes
Components of eukaryotic genes
Exons
Introns
transcription start site
promoters
enhancers
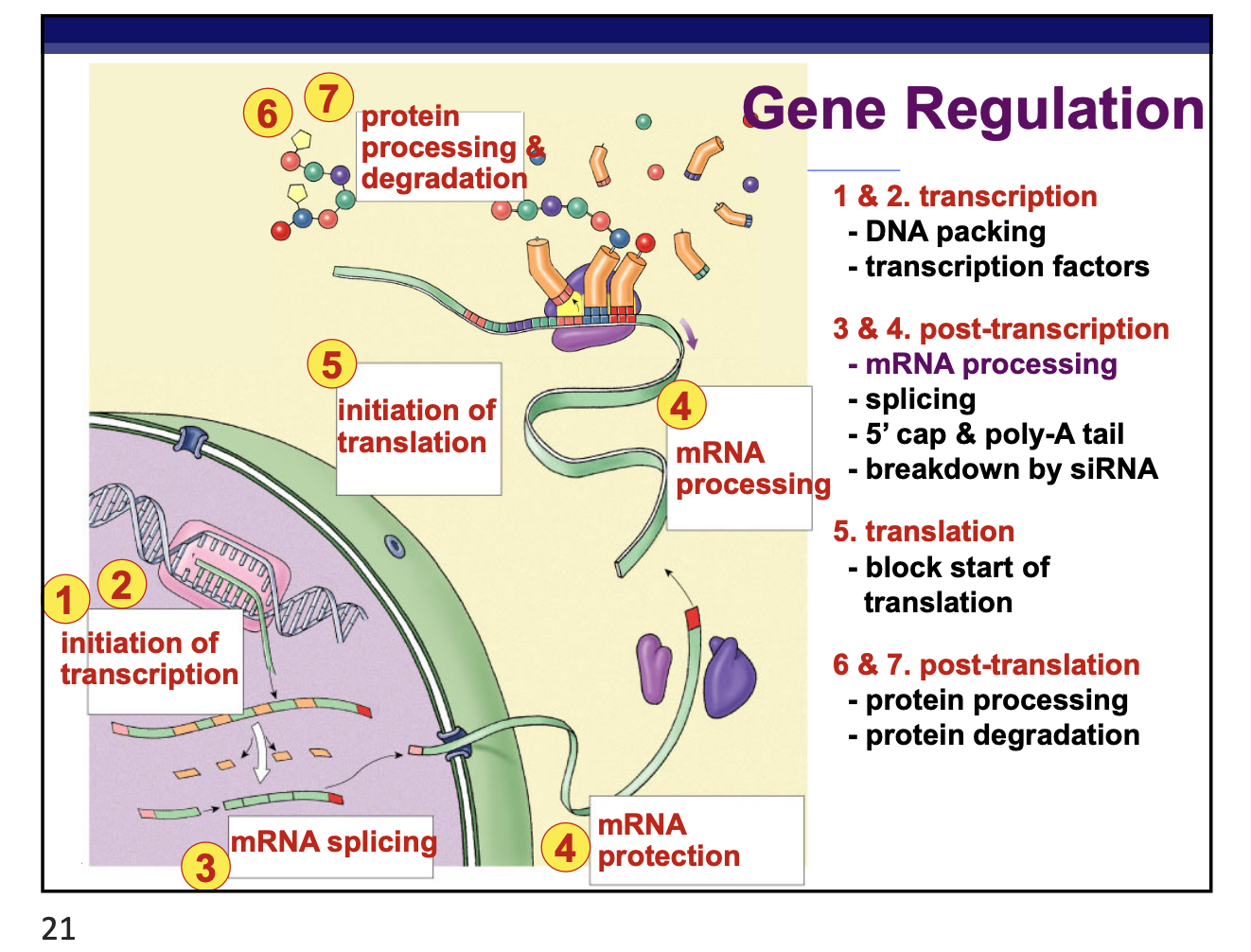
where can the control of gene expression occur
any step in the pathway from gene to functional protein
packing/unpacking DNA
transcription
mRNA processing
mRNA transport
translation
protein processing
protein degradation
What is DNA packing
DNA packing in gene control refers to how DNA is organized and condensed in the cell, affecting gene expression
Importance of DNA packing
looser packed DNA = higher transcription
transcription factors and RNA polymerase can bind to the gene promoter regions
Tightly packed DNA = lower transcription
prevents the binding of transcription factors and RNA polymerase to the gene promoter regions
negative gene regulation
signaling moleucles interact with repressor proteins that dictate whether a gene will be turned on or off
positive gene control
a signaling molecules generates a complex that interacts with the DNA
how can genes be unaccesible?
the way DNA is wrapped around histones play an important role in gene silencing
genes bound to histones can’t be expressed
to tightly packed
rna polymerase can’t access it
how can genes become accessible
Genes become accessible if histones undergo
acetylation
methylation
phosphorylation
This descreases the histones affinity for DNA
when a gene is no longer coiled around histone it becomes accesible
DNA methylation
methylation (addition of methyl group) of DNA blocks transcription factors
no transcription
genes turned off
nearly permanent
attachment of methyl group to cytosine
Histone acetylation
Acetylation of histones unwinds the DNA
enables transcription
making it accessible to RNA polymerase and transcription factors
genes turned on
attachment of acetyl group to histone
comfirmaional change in histone proteins (loose)
epigenetics
the effect of the environment on genes
epigenetic markers can turn genes on or off (eg. methylation and acetylation)
explains why identical twins can be different
can be inherited from earlier generations
transcription initiation (promoters)
control region on DNA / special region of DNA located near the start of a gene
It acts like a "signal" or "starting point" that tells the RNA polymerase + transcription factors where to begin transcribing a gene into RNA
“base rate” of transcription
transcription intitiation (enhancers)
control region on DNA that is located far from the gene (upstream or downstream)
activator proteins bind to it to increase the rate of transcription
Enhancers do this by bringing the promoter and transcription machinery closer together, even if they are far apart in the DNA sequence.
purpose of transcription factors
Transcription factors are proteins that regulate gene expression
transcription factors help recruit RNA polymerase and other general transcription factors to the gene’s promoter.
This creates a complex called the transcription initiation complex, which is necessary for the start of transcription.
purpose of transcription initiation complex
It ensures the accurate and efficient binding of RNA polymerase and transcription factors to the gene, allowing for proper gene expression. Without this complex, transcription cannot begin
How is the transcription initiation complex formed
Enhancers interact with proteins called activators
When activators bind to the enhancers, another protein can bend DNA to bring the activators closer to the promoter where the transcription factors can be found
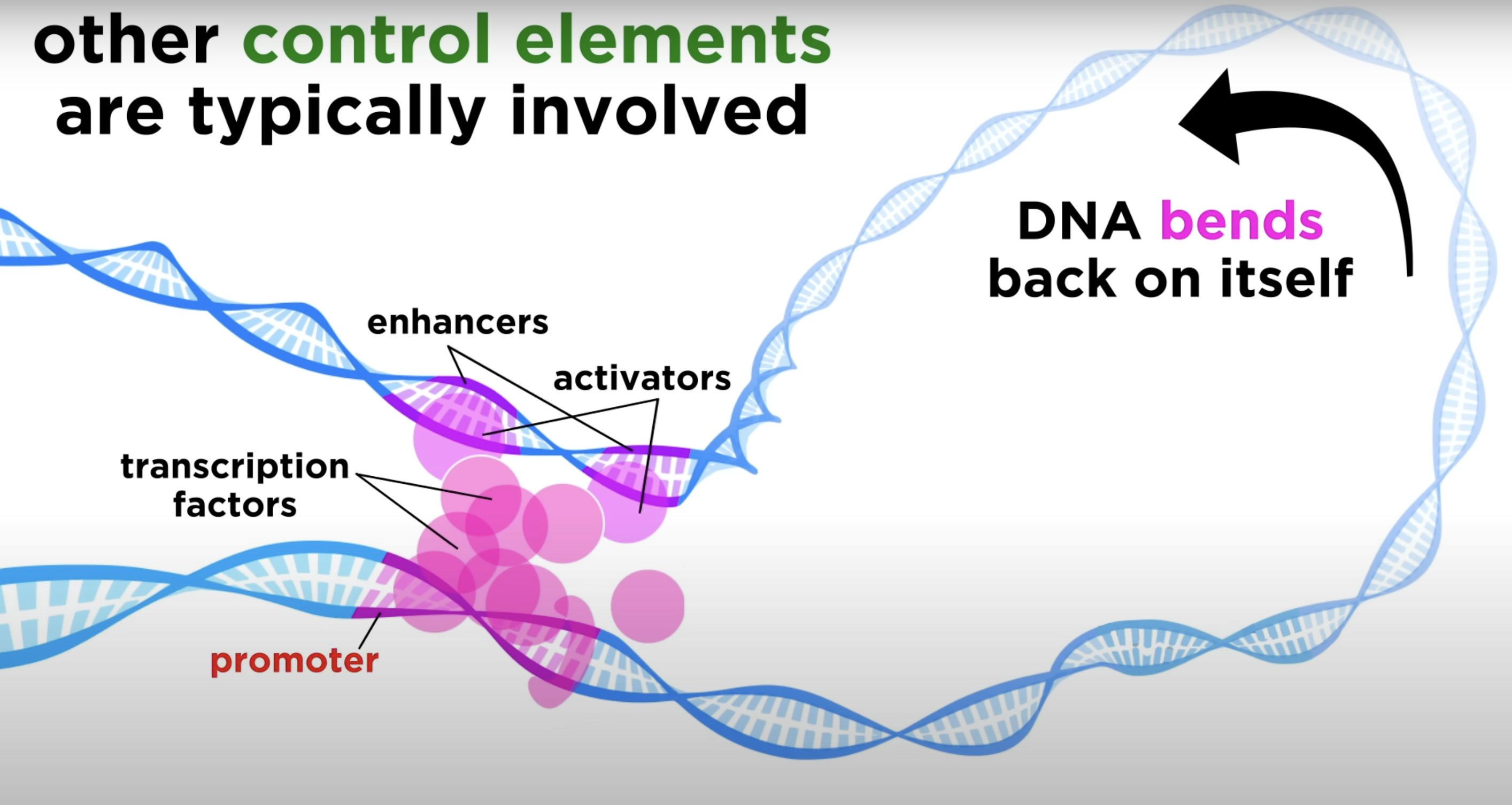
activator proteins
bind to enhancer sequence and stimulate transcription
can increase rate of transcription
silencer proteins
bind to enhancer sequence and block gene transcription
Regulation of mRNA degeneration
life span of mRNA determines amount of protein synthesis
RNA interference
small interfering RNAs (siRNA)
short segments of RNA used to silence genes by binding to and inducing the degradation of specific mRNAs
RNA interference is an example of ——
Post transcriptional control
turns off genes = no protein produced
how is siRNAs used in research
siRNAs are widely used in research to silence or knock down specific genes in order to study their function
siRNA-based therapies are being explored for the treatment of diseases such as cancer, viral infections, and genetic disorders, where the goal is to target and degrade the RNA of disease-causing genes
control of translation
post transcription regulation
Regulatory proteins attach to the 5’ end of mRNA and blocks initiation of translation stage
prevents attachment of ribosomal subunits and initiator tRNA
blocks translation of mRNA to protein
Protein processing
Protein processing refers to the modifications that occur after a protein is synthesized (translated) but before it becomes fully functional.
what does protein processing do
These modifications can activate the protein, enhance its function, or target it to specific cellular compartments.
Types of protein processing
folding
fold to correct 3D structure
cleaving
Many proteins undergo cleavage, where a portion of the protein is cut off. This is often essential for activation
adding phosphate groups
This modification can change the activity, function, or localization of a protein. Phosphorylation is a key regulatory mechanism in cell signaling
adding sugar groups targeting for transport
critical for protein stability, folding, and cell-cell recognition.
Protein degradation
Proteins must be continually degraded and removed to regulate protein levels
protein degredation examples (2)
ubiquitin tagging
protein is degraded
proteasome degredation
The proteasome breaks down the protein into small peptides, which are then degraded into amino acids and recycled
summarize gene regulation