Exam 4 2022 Study Set
1/61
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
62 Terms
Intermidiate filaments (tetramers)
Forms filaments that have no structural polarity - the two ends are the same
Microtubules (dimers)
Forms filaments that serve as tracks for molecular motors
Forms asymmetric filaments with distinct structural polarity
Actin (monomers)
Forms filaments that serve as tracks for molecular motors
Forms asymmetric filaments with distinct structural polarity
B on this microtubule is the what?
GTP binding site where GTP can be hydrolyzed and rapidly exchanged
C on this microtubule is the what?
GTP binding site where GTP cannot be easily exchanged
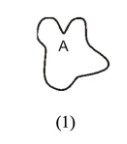
A on this actin is the what?
ATP binding site
![<p>At an [actin] of 0.4 μM, will there be net assembly or disassembly at the MINUS ends?</p>](https://knowt-user-attachments.s3.amazonaws.com/ff6858ce-8201-444a-87db-813a99c9f785.jpeg)
At an [actin] of 0.4 μM, will there be net assembly or disassembly at the MINUS ends?
disassembly
![<p>At an [actin] of 2.8 μM, will there be net assembly or disassembly at the PLUS ends?</p>](https://knowt-user-attachments.s3.amazonaws.com/2dce09df-baf1-4dcc-a5a7-8f81dfc32ac4.jpeg)
At an [actin] of 2.8 μM, will there be net assembly or disassembly at the PLUS ends?
assembly
![<p>At an [actin] of 0.4 μM, will there be net assembly or disassembly at the PLUS ends?</p>](https://knowt-user-attachments.s3.amazonaws.com/d380c5f1-1811-4dda-a95c-1a67e420a7c2.jpeg)
At an [actin] of 0.4 μM, will there be net assembly or disassembly at the PLUS ends?
disassembly
![<p>At an [actin] of 2.8 μM, will there be net assembly or disassembly at the MINUS ends?</p>](https://knowt-user-attachments.s3.amazonaws.com/6fa4f1ef-94e5-42ab-8ab2-b9730e9bbbf0.jpeg)
At an [actin] of 2.8 μM, will there be net assembly or disassembly at the MINUS ends?
assembly
![<p><span>How will these actin filaments behave at an [actin] of 1.5 uM? Be specific about what happens at each end and use the name of this effect.</span></p>](https://knowt-user-attachments.s3.amazonaws.com/987a7775-59f0-4294-aa5c-4a22290973cb.jpeg)
How will these actin filaments behave at an [actin] of 1.5 uM? Be specific about what happens at each end and use the name of this effect.
Actin subunits will add to the filament plus ends and come off of the filament minus ends. Thus monomers will process through the filaments from the plus to the minus end, a state called “treadmilling.”
Actin nucleotide-exchange proteins such as profilin are responsible for…
At the front of lamellipodia, polymerization-competent actin monomers are abundant
Actin plus-end capping proteins are responsible for…
Lamellipodia have a broad front, supported by an extensively branched network of short actin filaments
Actin minus-end capping proteins are responsible for…
At some times in the cell, actin filament dynamics reflect only the properties of the filament plus end
Actin side-binding proteins are responsible for…
Actin filaments in muscle sarcomeres are both highly stable and have their myosin-binding sites blocked at rest
Proteins such as cofilin that sever actin filaments in regions rich in ADP-actin subunits are responsible for…
At the rear of lamellipodia, actin filaments
Actin binding proteins such as spectrin, with two actin binding sites that are far apart are responsible for…
Many cells have loose meshworks of actin in which the filaments intersect each other at different angles
Motor proteins such as myosin V are responsible for…
Organelles can translocate along actin filaments toward their plus ends
Actin binding proteins such as fimbrin, which have two actin binding sites that are very close together are responsible for…
Microvilli can resist shearing forces in the gut because they are supported by a core of closely-spaced, linked actin filaments
Regulated monomer-sequestering proteins are responsible for…
In platelets, a signal can generate a burst of actin polymerization without the need to synthesize additional actin
ARP complex, a protein that binds the walls of actin filaments and nucleates the assembly of new filaments are responsible for…
Lamellipodia have a broad front, supported by an extensively branched network of short actin filaments.
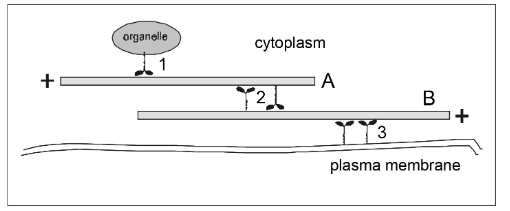
If filament A has a fixed position, then in which direction will the organelle go when the motor protein at (1) operates?
motors = dynein
motors = myosin
right
left
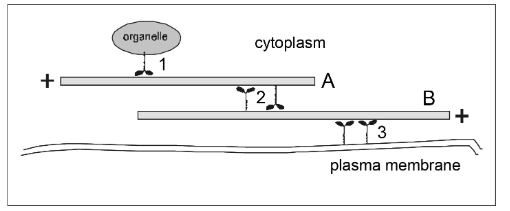
If motor protein (3) operates, then in which direction will filament B move?
motors = dynein
motors = myosin
right
left
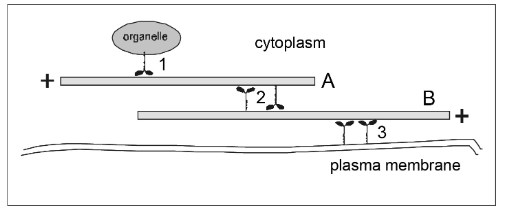
If filament B has fixed position, in which direction will filament A move if the motors at (2) operate?
motors = dynein
motors = myosin
left
right
Order from largest (1) to smallest (5)
an intestinal epithelial cell
a microvillus
Maturation Promotion Factor (MPF)
cyclin protein
ATP
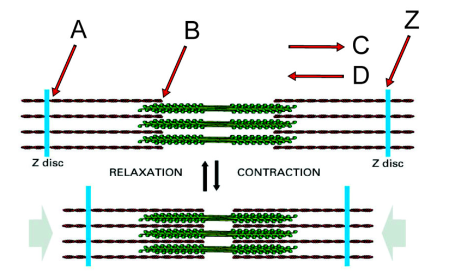
Direction in which the myosin heads on the right side of the structure will translocate along actin filaments during contraction
C
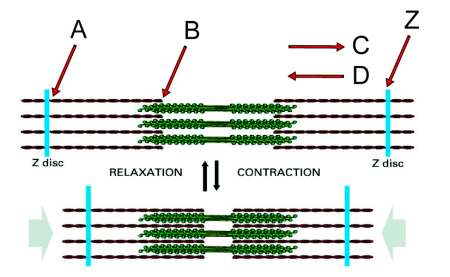
Minus end of an actin filament
B
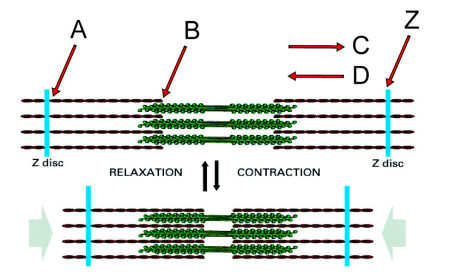
Plus end of an actin filament
A
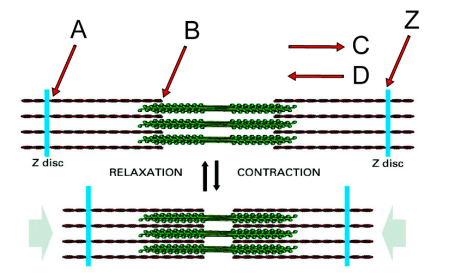
If you anchored the Z disc on the left, in which direction would the Z disc labeled “Z” move during myosin activity?
Left
What second messenger regulates the interaction of myosin heads with the actin filaments?
Calcium
What proteins mediate the effect of this second messenger?
The troponin complex and tropomyosin
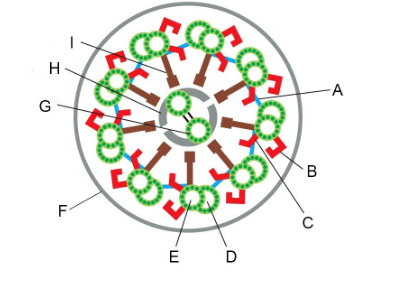
Structures that can be removed by the salt extraction of axonemes, leaving most of the structure intact but eliminating beating (two)
B and C (outer and inner dynein arms)
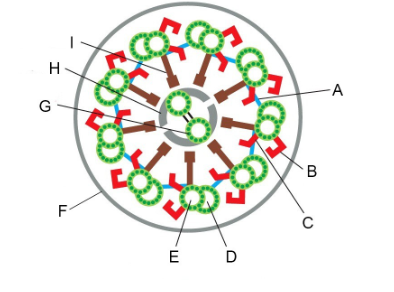
Structure that can be removed by detergent, allowing you to add ATP and get beating of the axoneme in vitro
F (plasma membrane)
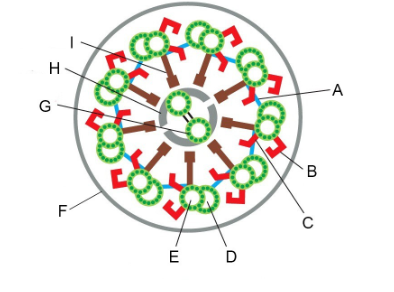
Structure with which dynein motor proteins interact cyclically when they generate force
D (B microtubule)
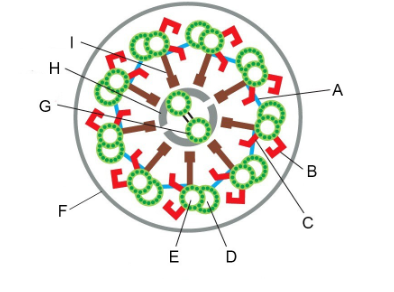
Structures that constrain sliding forces into ciliary bending (two)
A and I (linking protein/nexin links, radial spoke/central spoke)
We looked at a key experiment that lead to our current understanding of how flagella beat, the “sliding disintegration” experiment. Summers and Gibbons prepared sea urchin sperm flagella by stripping off the membranes with detergent. If they added ATP to these flagellar axonemes, they could beat. But if they treated them briefly with the protease trypsin and then added ATP, instead of beating, the axonemes would slide apart. What did the protease treatment do to convert the flagellar beating into simple sliding? Be specific with regard to two things that happened.
It cut the links between the doublet MTs, specifically the nexin links (A, above) between doublets and the central spokes (I, above) that link the outer doublet MTs to the central pair.
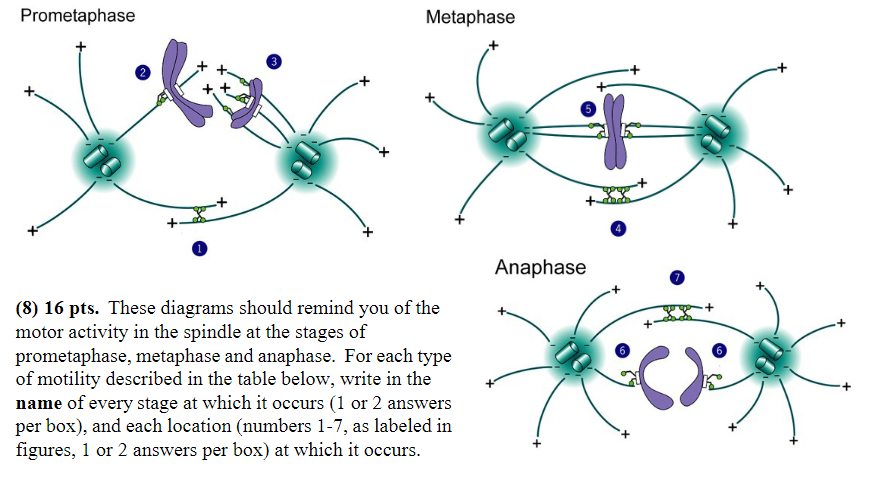
Minus-end-directed motor proteins exert force that successfully moves chromosomes:
Stage: prometaphase, anaphase
Location: 2, 6
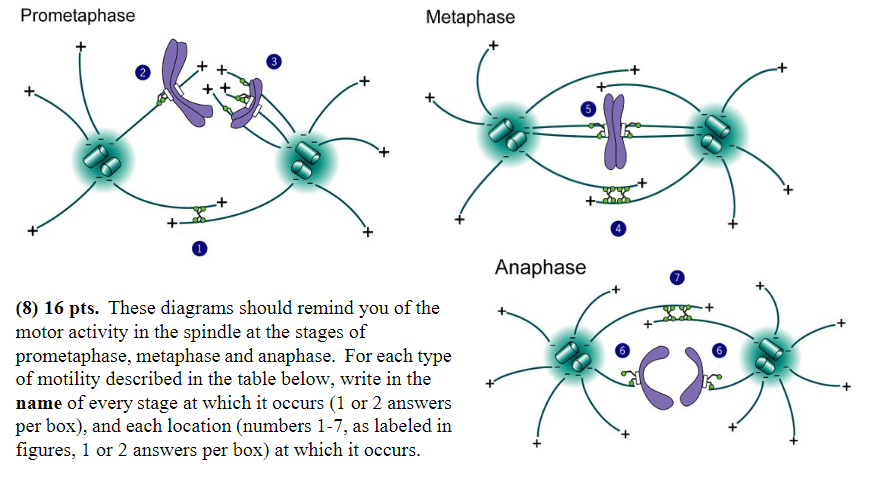
Minus-end-directed motor proteins exert force that “tries” unsuccessfully to pull the spindle poles together:
Stage: metaphase
Location: 5
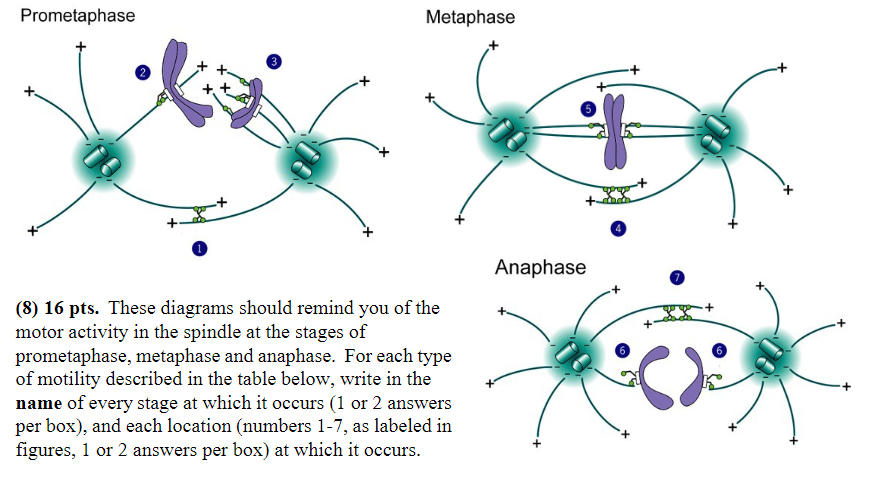
MT disassembly is thought to generate or regulate movement:
Stage: anaphase
Location: 6
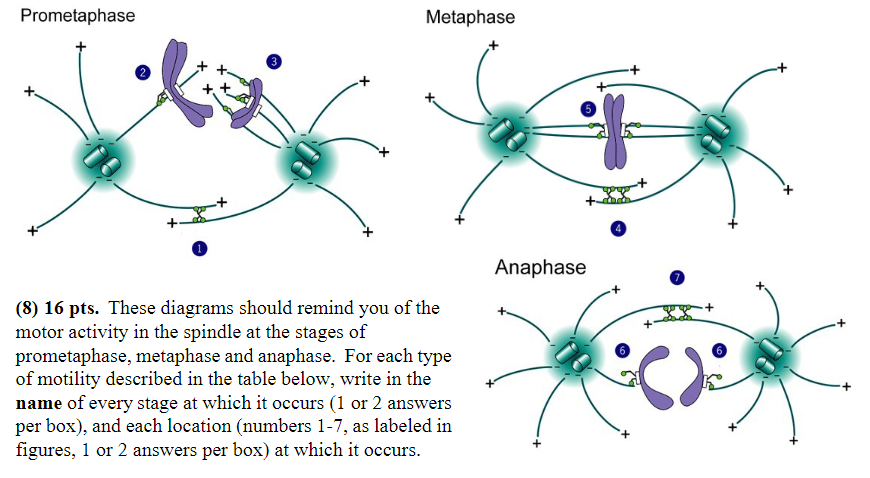
Plus-end directed motor proteins exert force that moves chromosomes:
Stage: prometaphase
Location: 3
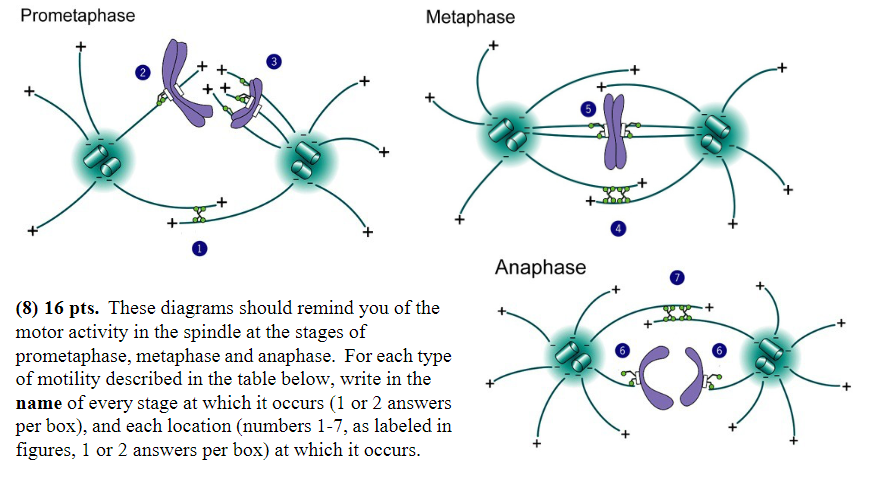
Plus-end-directed motor proteins exert force that “tries” unsuccessfully to push the spindle poles apart:
Stage: metaphase
Location: 4
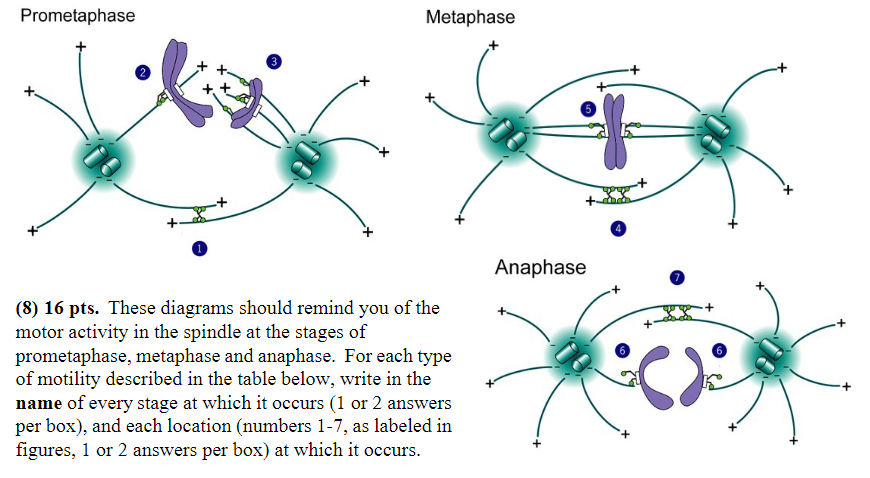
Plus-end-directed motor proteins exert force that successfully pushes the spindle poles apart:
Stage: prometaphase, anaphase
Location: 1, 7
At anaphase, what is the behavior of the plus ends of the polar MTs?
assembly [also recruitment of new motors]
At anaphase, what is the behavior of the ends of the kinetochore MTs?
Plus end: disassembly
Minus End: Capped (no change)
One of the most interesting transitions in mitosis is the change in the kinetochore MTs as the cell enters anaphase. How does their status (behavior) change at this transition? Please be certain to include what they are doing prior to entry (in metaphase) and after entry (in anaphase). That is the only way to clearly indicate the change
They stop treadmilling and shift to plus-end disassembly
How does the motor protein activity associated with the kinetochore MTs change at the transition from prometaphase to metaphase?
The plus-end directed motor activity is lost
Cells of this strain getting larger with each round of division and dying out after several cell cycles results from…
It pauses at the G2/M checkpoint until the cell volume has increased 3-fold since the last cell division
Cells of this strain getting smaller and smaller with each round of division and dying after several cell cycles results from…
It crosses the G2/M cell cycle checkpoint before the cell has doubled in volume since the last cytokinesis
The protein phosphatase that normally removes the inhibitory phosphate (the one that was put on by Wee1) from the mitotic cell cycle kinase is mutated and constitutively active (on all the time) in this strain
Cells of this strain do not divide. They grow in size and completely duplicate their DNA, but they can’t enter M – they are arrested in G2 and do not even undergo nuclear envelope breakdown. This results from…
The cyclin for the G2/M checkpoint is degraded prematurely and remains at an almost undetectable concentration in the cell
The G2/M cyclin cannot bind to the cell cycle kinase
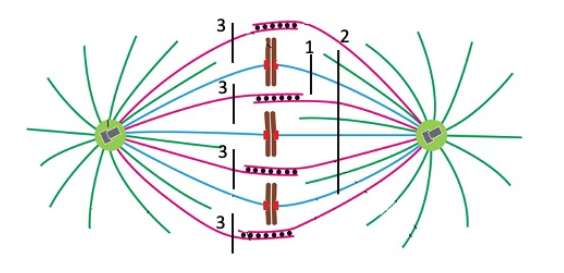
The spindle poles would be pulled toward each other.
Cut 3
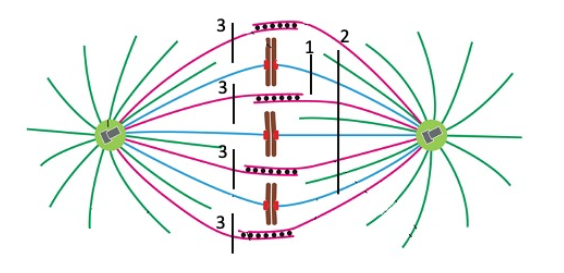
All chromosomes would begin to move toward pole at left, and the poles would probably begin to separate.
Cut 2
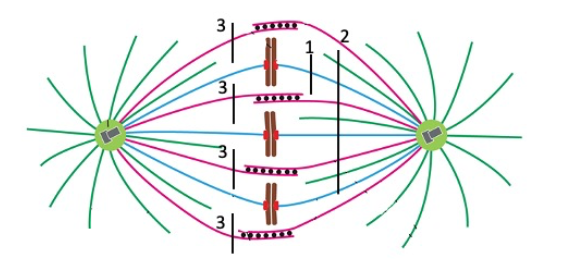
One chromosome would move toward the pole at right.
none
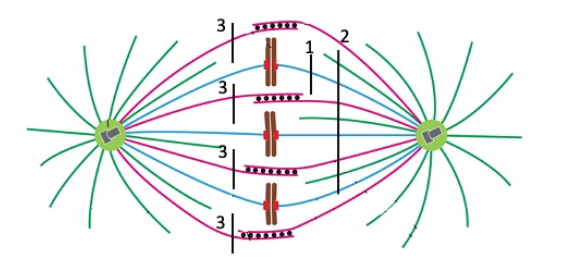
One chromosome would move toward the pole at left.
Cut 1
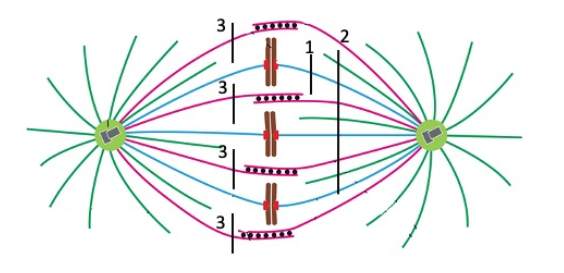
The spindle poles would begin to separate
None (or cut 2, unless I change the question to ONLY spindle poles would seperate)
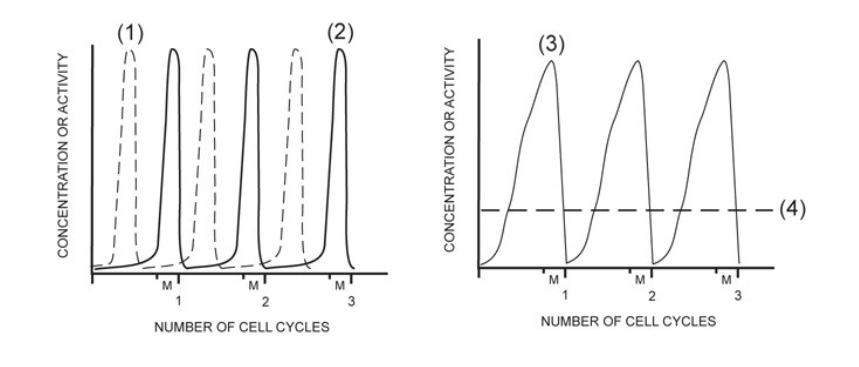
activity of MPF
2
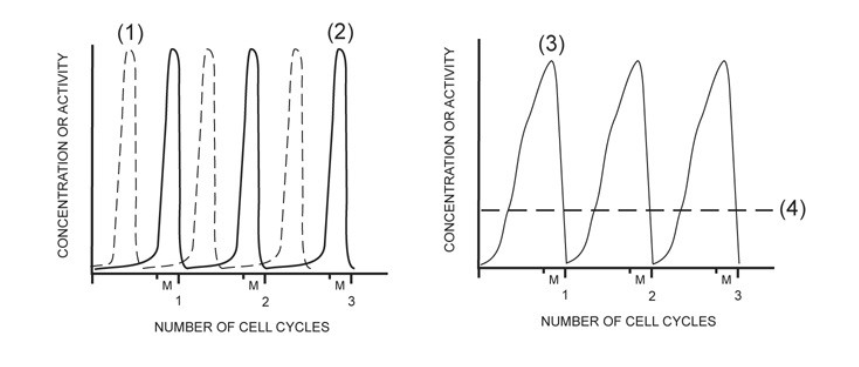
concentration of G1/S cell cycle kinase (S-Cdk)
4
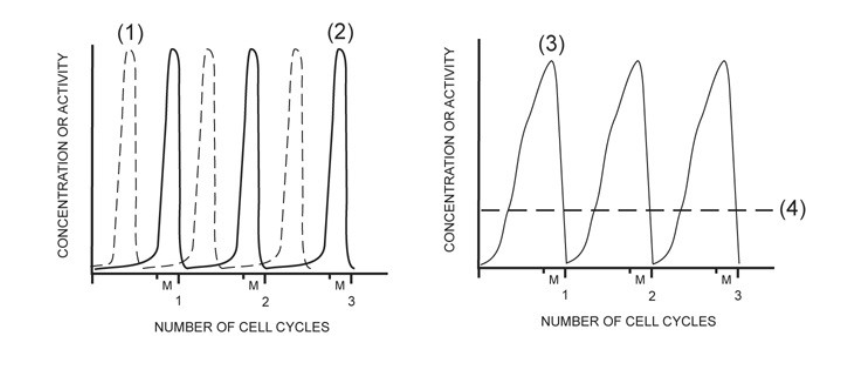
activity of G1/S cell cycle kinase (S-Cdk)
1
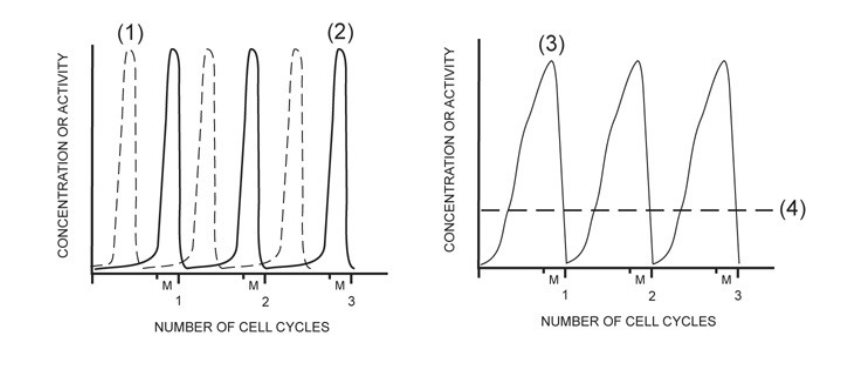
concentration of mitotic cyclin (M-cyclin)
3
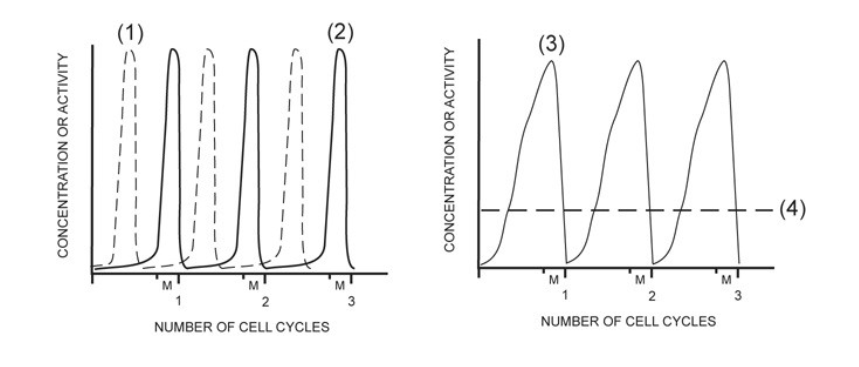
concentration of S-cyclin (for the G1/S checkpoint)
NONE
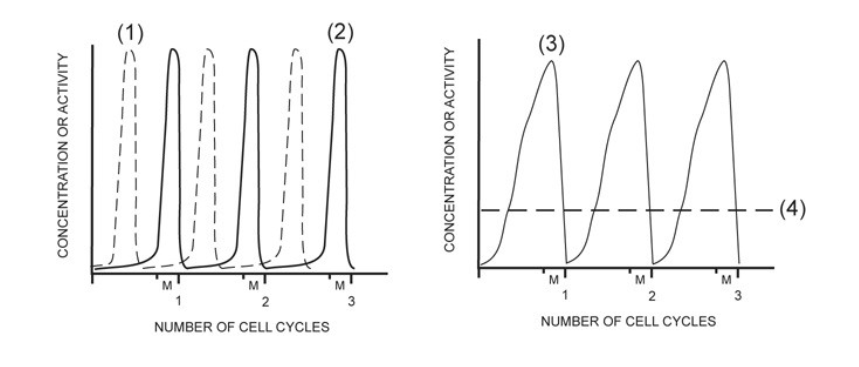
concentration of mitotic cell cycle kinase (M-Cdk)
4
In one sentence, describe the exact composition of MPF. You must include all the essential components, and their state:
It was the mitotic Cdk (kinase), bound to mitotic cyclin, and modified post-translationally by phosphorylation at a stimulatory site
In the cell fusion experiments, Rao and Johnson found that when they fused a G1 phase cell with an S phase cell, the G1 nucleus was forced ahead into replicating it’s DNA. What happened when they fused an S phase cell with a cell in G2, how did the G2 nucleus respond?
The G2 nucleus was no longer able to respond and did not enter (inappropriate) DNA synthesis