Introduction to bacterial genetics
1/55
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
56 Terms
Microbial genetics
Is the study of the inheritance of the characteristics of a microbial cell and how these characteristics can vary
all phenotypes are encoded in the genome - including virulence factors, what media it grown on, lab tests
Is used to engineer strains of microbes which have desirable characteristics - bacteria is often used in industrial fermentation to produce useful products. below is example of bacteria cell being engineered to produce human insulin gene - easy to grow to large levels due to fast replication
Helps us understand how microbes become resistant to antimicrobial agents and how to prevent such resistance emerging
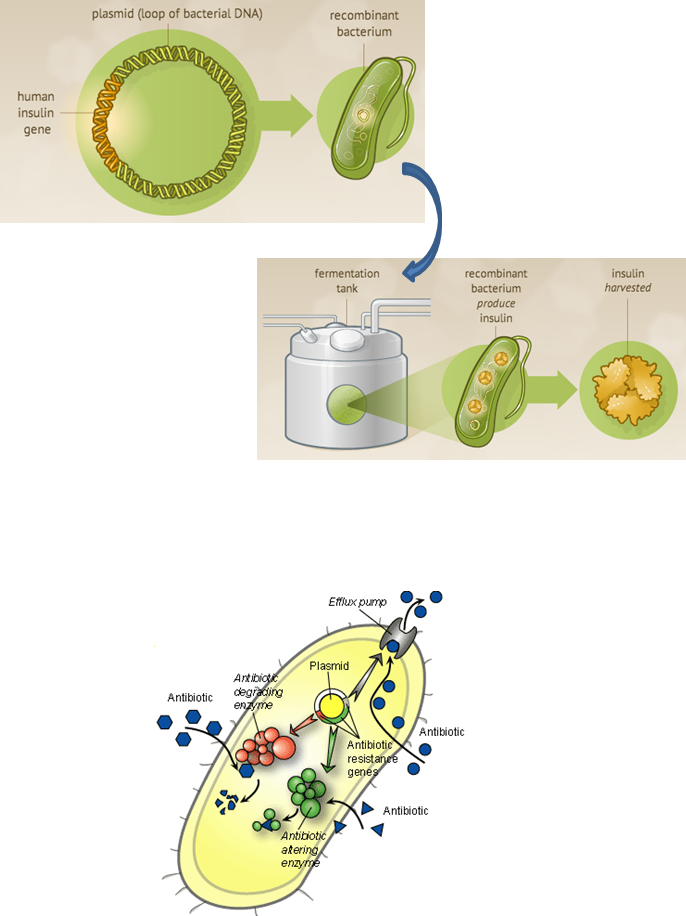
The genetic material of bacteria
Exists in the BACTERIAL CHROMOSOME which is separate from the cytoplasm but not bound by a nuclear membrane
Exists in PLASMIDS which are transient circular genetic elements in the cytoplasm - extra chromosomes which are a lot smaller and don’t usually carry essential genes but instead beneficial genes e.g. genes helping with ABR, or a gene allowing virulence
DNA
Chemically, genetic information is carried by the nucleic acids: DNA and RNA (ribonucleic acid)
RNA is the intermediate molecule that converts RNA and genetic information to amino acids
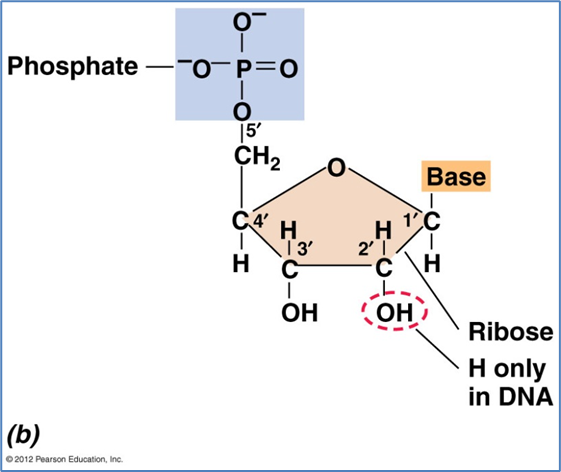
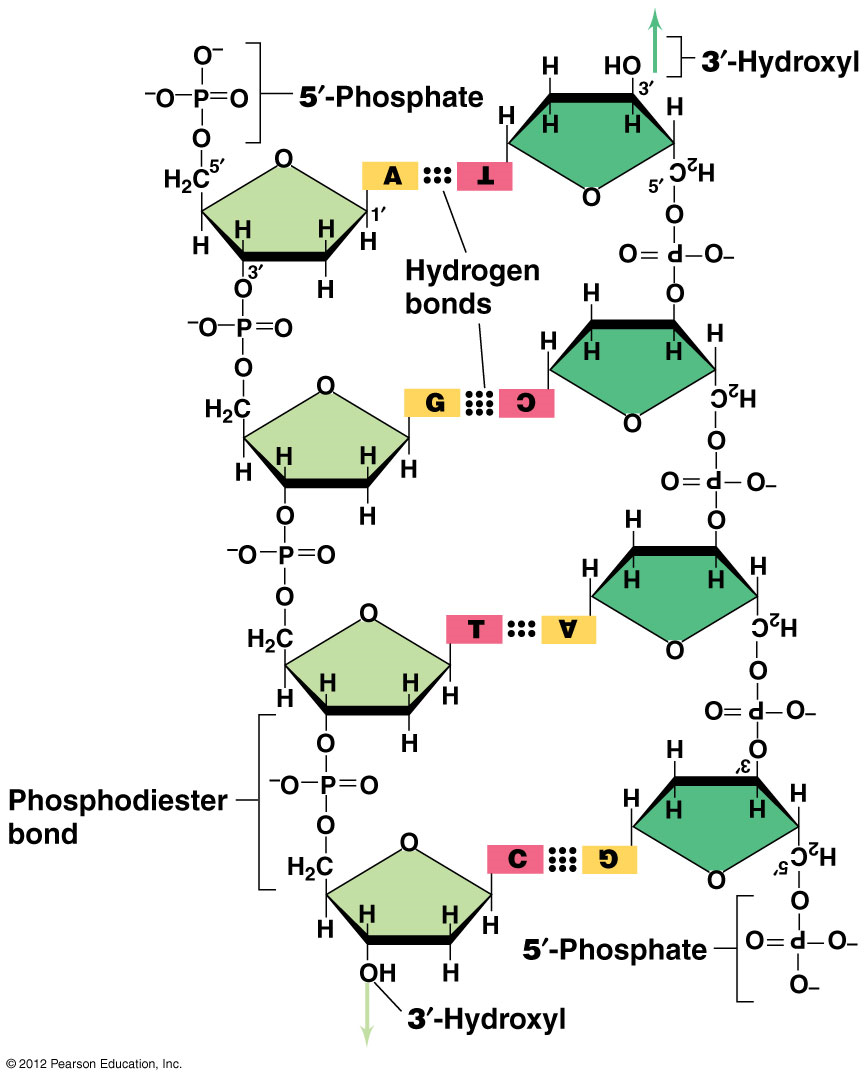
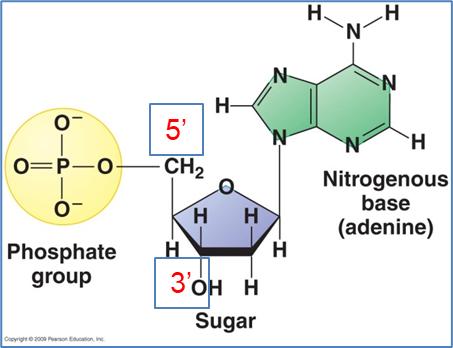
The monomers of nucleic acids (DNA and RNA) are nucleotides:
Pentose sugar – ribose or 2’deoxyribose
Nitrogen base - ADTG
Molecule of phosphate (PO43-) - sugar phosphate backbone
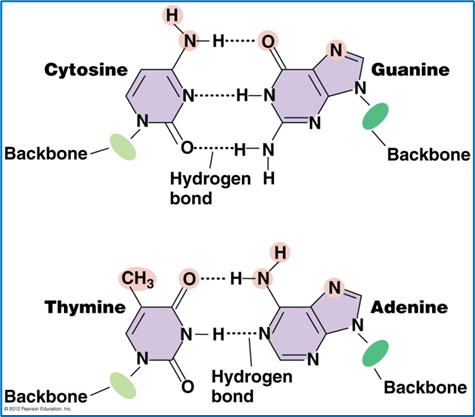
Bases complement each other:
Purine with pyrimidine
Adenine – Thymine - 2 H BONDS
Guanine – Cytosine - 3 H BONDS
This sequence of bases represents the organism’s genetic code
The two sugar-phosphate chains are antiparallel (5’ to 3’ and 3’ to 5’) and are twisted in a double helix
Each twist of the helix is 10 base pairs
The structure is supercoiled
bases are attached to C1
Phosphate is attached to 5 prime Carbon-
Diagram
Replication of the Bacterial Chromosome
Occurs in a semi-conservative manner and is usually bidirectional
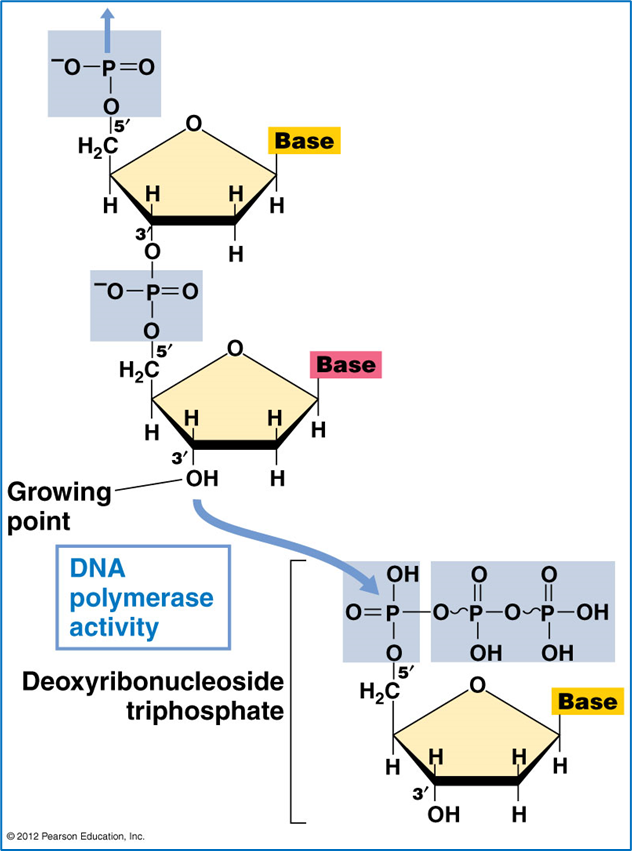
The precursor of each new nucleotide in the DNA strand is a deoxynucleoside 5’triphosphate
Nucleotides are added to the new strand by DNA polymerase only in the 5’-3’ end (leading strand)
In the 3’-5’ direction, synthesis is discontinuous in short Okazaki fragments using an RNA primer. Fragments then joined together with DNA ligase (lagging strand)
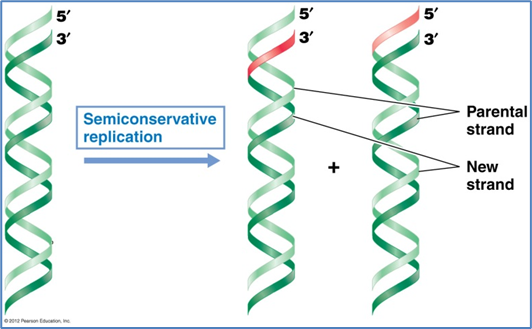
Helix s unwound and a new strand is synthesised complementary to the parent strand which is the template - semi conservative as there is 1 parent strand and 1 new strand
Bi-directional so very efficient e.g. E.coli can replicate in 20 minutes
Nucleotides can only be added to the new strand by DNA polymerase in the 5 to 3 prime
In other strand since it is going in other direction we need Okazaki fragment - lagging or discontinuous strand
Semi conservative replication
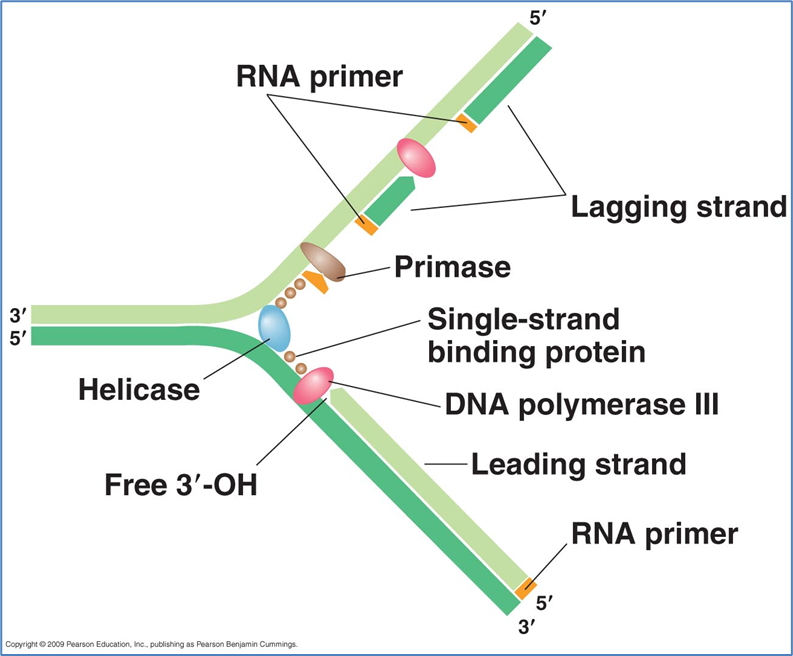
Replication fork - two strands split to allow synthesis of new strands of DNA
On other strand small RNA primers are synthesised along the length of the strand so that the DNA can add the nucleotide in the 5 to 3 prime direction using DNA polymerase
to do this two terminal phosphates are removed from the new nucleotide and inner phosphate is attached covalently to the deoxyribose of the growing chain at the three prime carbo. this relies on the free hydroxyl of the three prime carbon - see picture 1 for reference
diagram
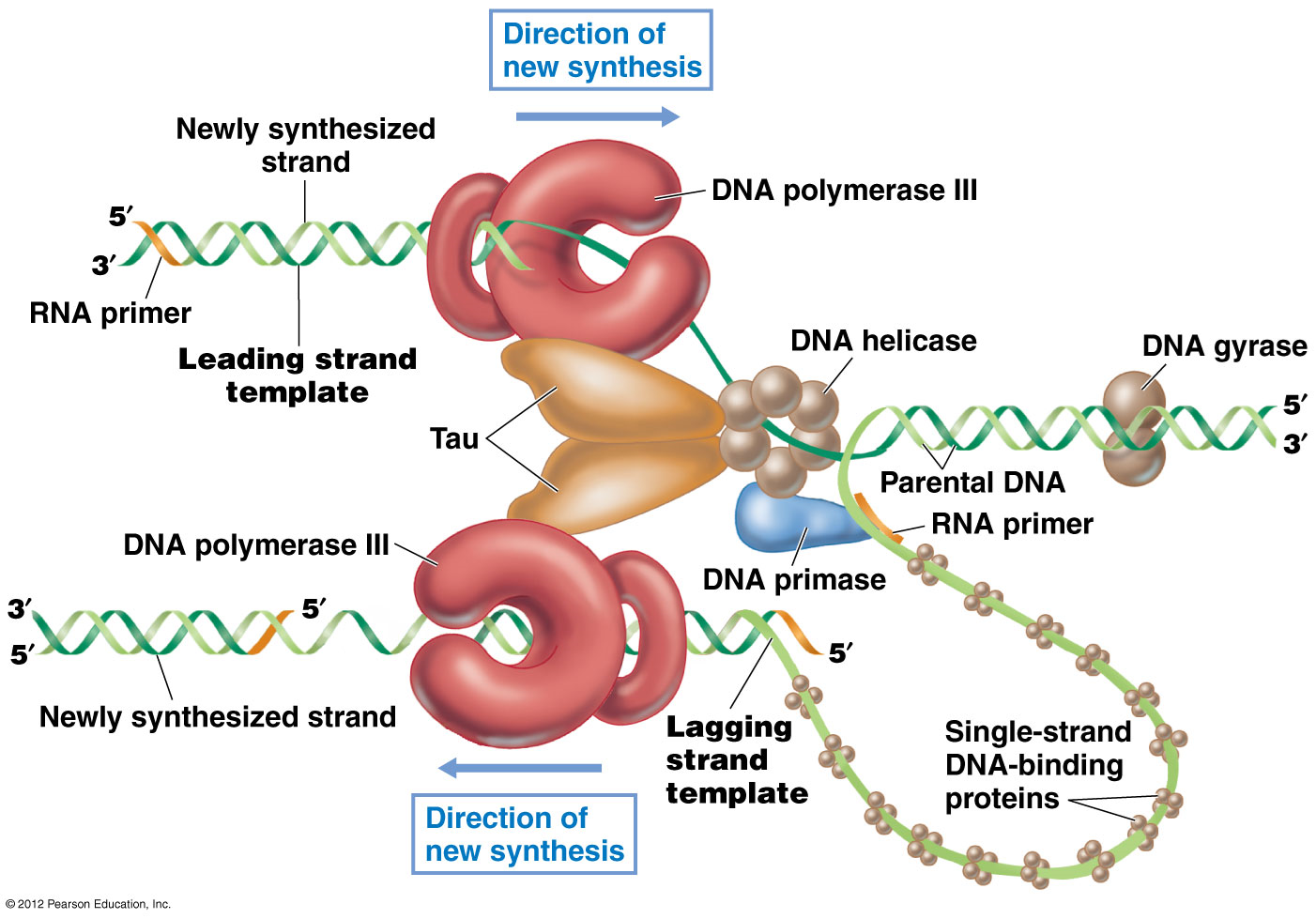
small RNA primer is attached at the beginning and then the DNA polymerase adds nucleotides
on the lagging strand it is discontinuous. Small RNA primers are synthesised by the DNA primase enzyme - normally 11 base pairs long
DNA polymerase can then add nucleotides from each of these primers in 5 to 3 direction - stops when it reaches previously synthesised DNA
after this the primer is removed and is replaced with more nucleotides - these are called okazaki fragments
DNA ligase seals the end gaps between the fragments
TAU unit is there to stabilise the complex
DNA helicase is responsible for unwinding the supercoiled DNA
DNA gyrase prevents unwanted supercoil occurring
DNA binding proteins also stabilise the structure
Replication of circular DNA
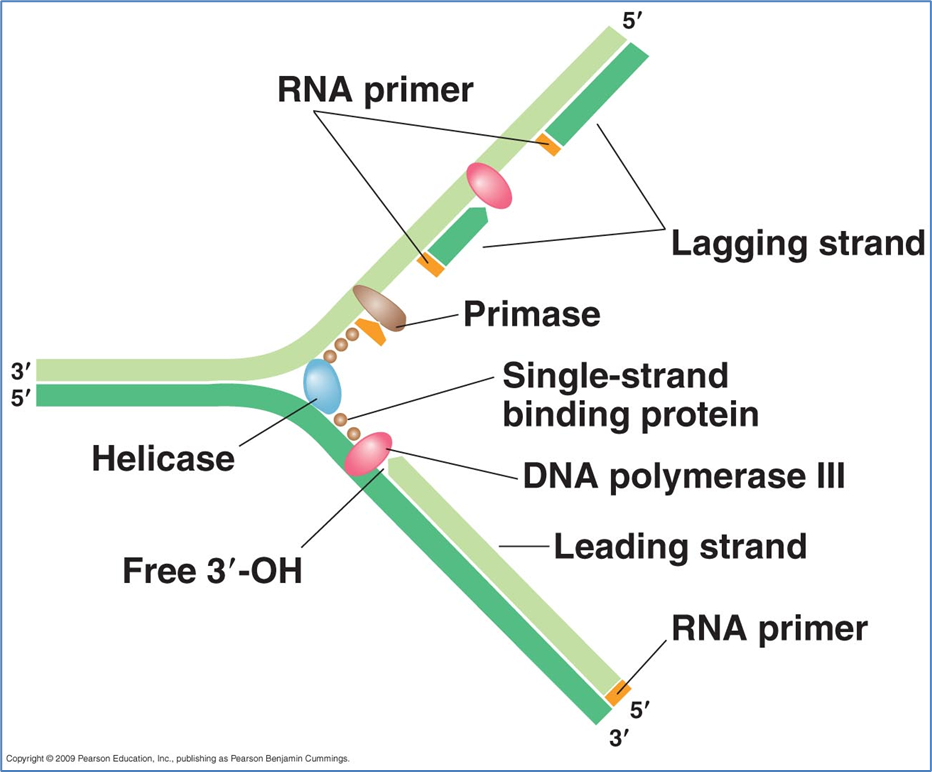

Map of the Chromosome of E. coli K-12
4,639,221 base pairs
4288 open reading frames (indicate genes
a gene is the unit of genetic information and the genome is the cells complete set of genes
many of the genes which encode enzymes of a single biochemical pathway are often clustered into what we call operands
an operand is a set of adjacent genes which is under the control of one promoter and the mRNA is then synthesised in one piece
for example if the bacteria needed to make some triptonan it would transcribe this operand to produce mRNA and then that would then produce the proteins for each of the five proteins for tryptophan biosynthesis - makes it more efficient as they are all clustered together
40 % of the predictive proteins in E. Coli are still unknown
Transcription
Sections (corresponding to genes) of ONE strand of the DNA molecule are transcribed
This is the process of converting DNA into mRNA and is carried out by RNA polymerase - RIFAMPCIN targets RNA polymerase
There is an initiation site (promoter) and termination site, which are specific nucleotide sequences - promoter region is found upstream while termination site is at the end of the gene where synthesis stops
The RNA polymerase moves down the DNA chain, temporarily opening the double helix and transcribing one of the strands
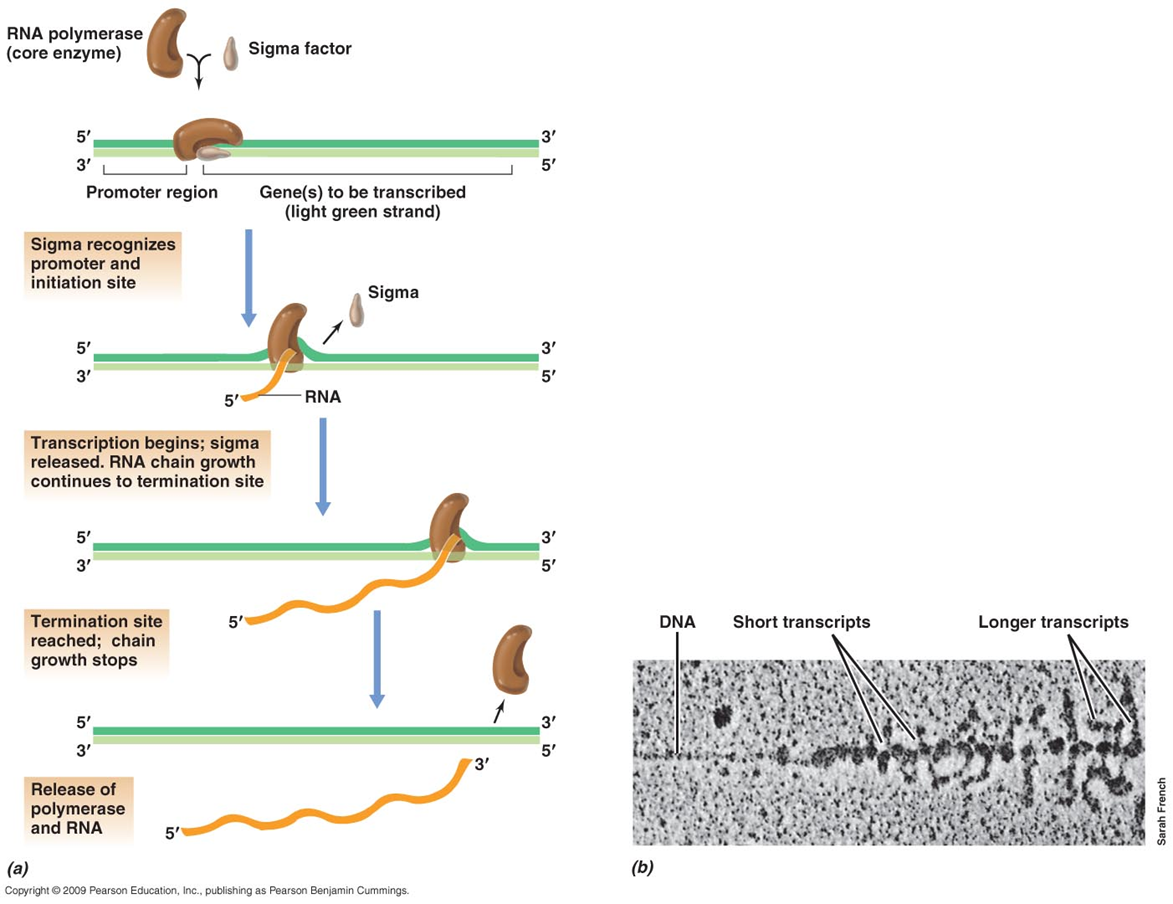
RNA polymerase must bind to sigma factor before it can work as it produces the holo enzyme which can then bind to the promoter region on the DNA
it then moves along the DNA and as it does that RNA is transcribed to MRNA - Sigma factor can dissociate
it then reaches the termination site and the enzyme is released and can bind to another sigma factor.
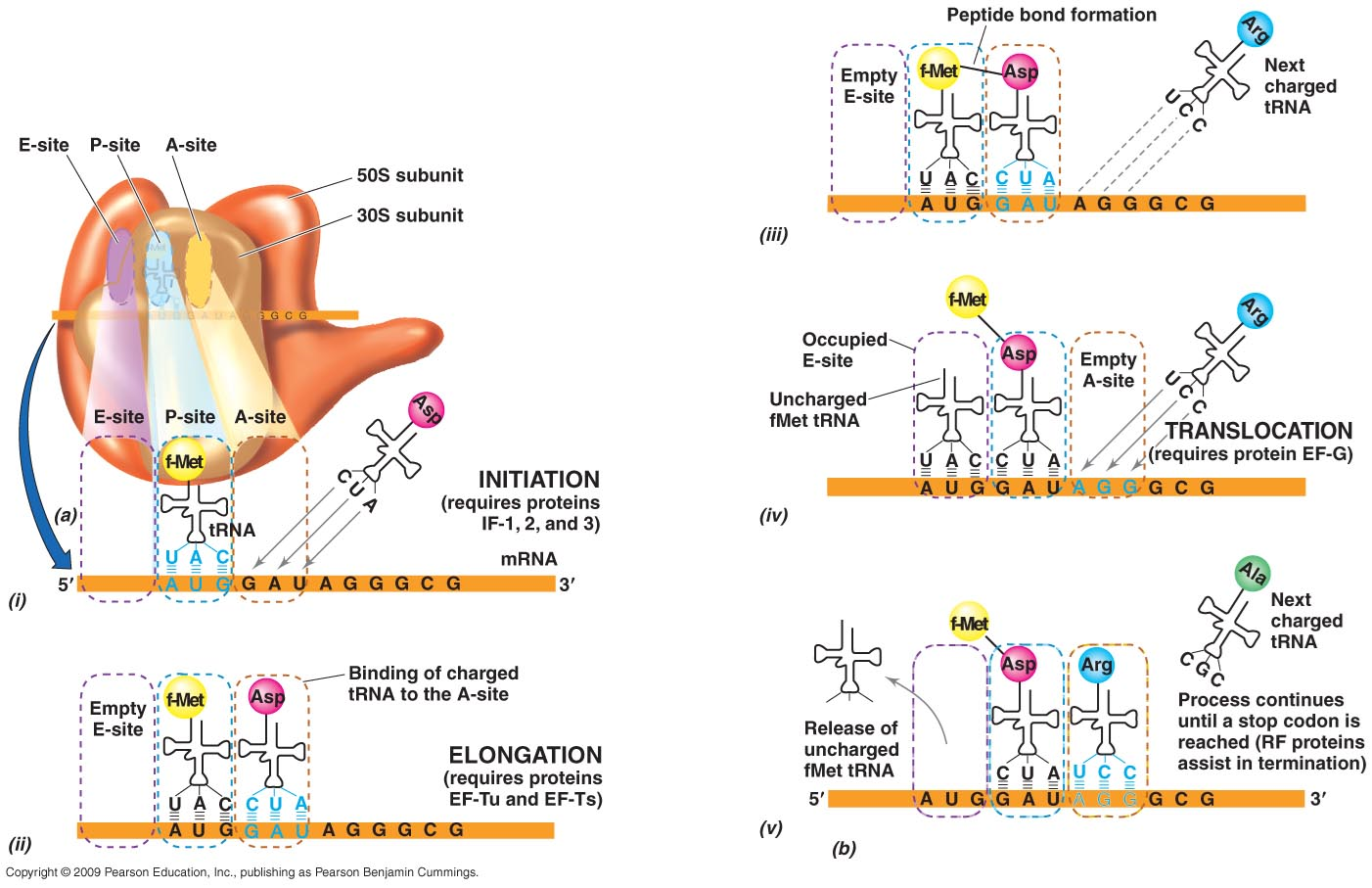
Translation
The mRNA has triplet sequences of bases – codons
Codons encode the 20 amino acids
Some codons act as start codons (AUG - methionine) and stop codons (UAA, UAG, UGA) which regulate the process of translation
After transcription, the newly-formed mRNA binds to a 30-S subunit of a ribosome - good AB target
Each amino acid associates with a molecule of tRNA. This tRNA contains a triplet base sequence (anticodon) which is complementary to the codons on the mRNA
The anticodon of tRNA for the first amino acid (usually a methionine) binds to its complementary mRNA codon at the appropriate place on the ribosome
A 50-S subunit associates with the 30-S subunit to form a complete working ribosome
A 2nd amino acid and its tRNA is added at the appropriate place on the ribosome and a peptide bond forms between the 2 amino acids
The first tRNA molecule then dissociates from the ribosome and can pick up another molecule of its specific amino acid
The ribosome then moves along the mRNA to the next codon and the next tRNA-amino acid complex is added
Growth of the polypeptide chain stops when a stop codon is reached
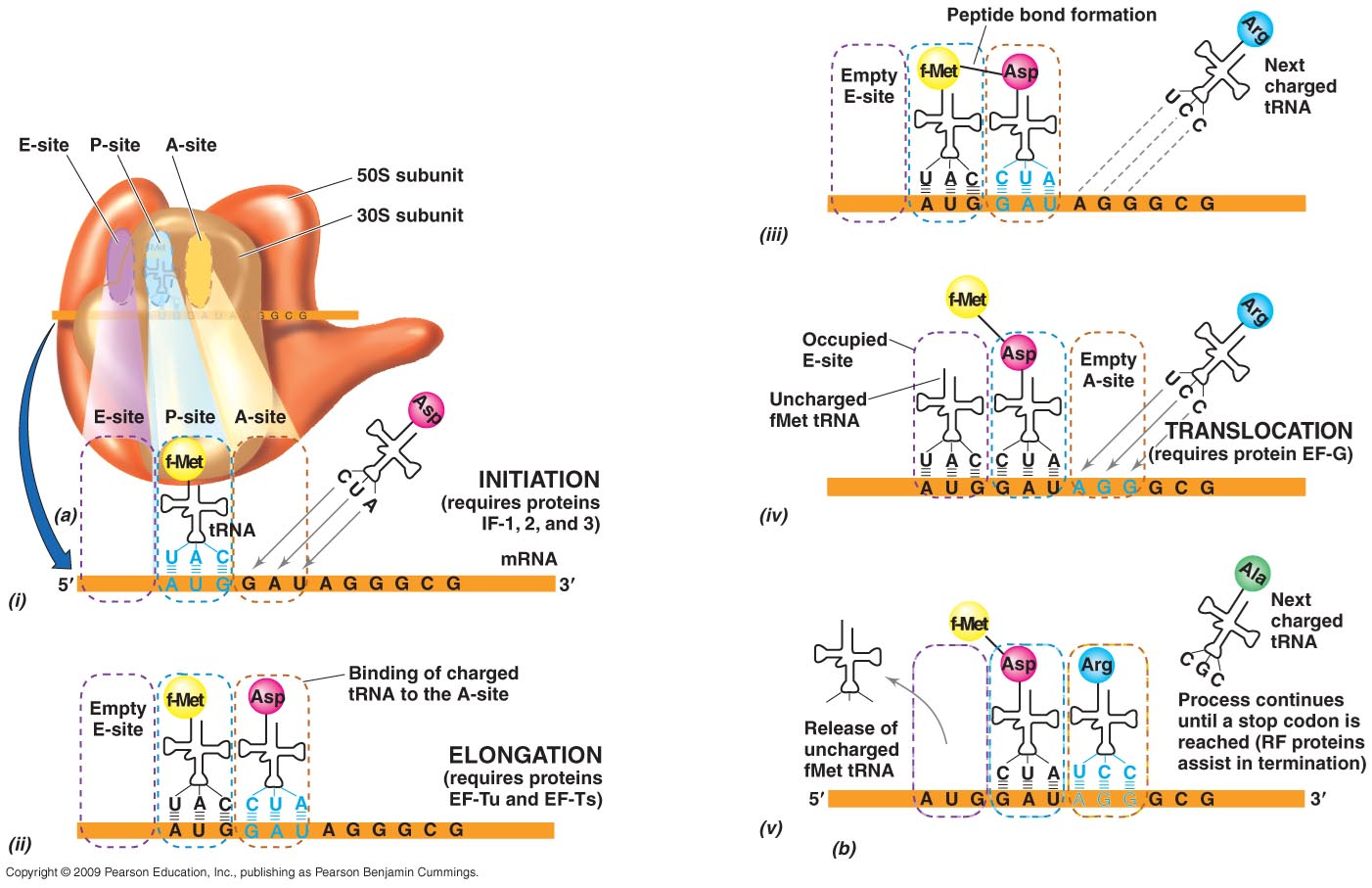
1st stage is that the 502 subunit associates with 302 subunit to form a complete working ribosome
second amino acid moves in and its trna is added to the mrna and a peptide bond can form between them
third amino acid - trn
Plasmids
Genetic elements that replicate independently of the host chromosome
Replicates via semi-conservative replication - does so independently to the chromosomal DNA
Small circular molecules
Range in size from 1 kbp to >1 Mbp
Carry a variety of non-essential, but often very helpful, genes
Can carry genes for AB resistance and virulence factors
Their abundance in the cell is variable
Genotype and phenotype
Genotype
The entire array of genes in the organism, i.e. the cell’s total genetic potential
Phenotype
The characteristics expressed by the cell under a particular set of conditions
Examples of Phenotypes Conferred by Plasmids in Prokaryotes
Phenotype Class | Organism |
Resistance | |
•Antibiotic resistance | Wide range of bacteria |
•Resistance to toxic metals | Wide range of bacteria |
Virulence | |
•Bacteriocin production and resistance | Wide range of bacteria |
•Animal cell invasion | Salmonella, Shigella, Yersinia |
•Coagulase, haemolysin, enterotoxin | Staphylococcus |
•Toxins and capsule | Bacillus anthracis |
•Enterotoxin, K antigen | Escherichia |
AB resistance - some bacteria are able to produce beta lactamases which breaks down beta lactam ring found in penicillin’s and cephalosporins
Mutation and mutants
Mutation
Heritable change in DNA sequence that can lead to change in the phenotype
Mutant
A strain of any cell or virus differing from parental strain in the genotype
Wild-type strain
Typically refers to a strain isolated from nature
Can be good or bad
beneficial mutation example - bacterial cell showing increased tolerance to an AB
Isolation of mutants
Selectable mutations
Give the mutants a growth advantage under certain environmental conditions
Useful in genetic research
Non-selectable mutations
Have neither an advantage or disadvantage over the parent
Detection of such mutants requires screening a large number of colonies and looking for the difference
e.g. ability of bacteria to produce a pigment
Molecular basis of mutations
Induced mutations
Made deliberately by using mutagens. Mutagens can be chemical, physical or biological. Examples is X RAY or UV.
Spontaneous mutations
Occur as a result of exposure to natural radiation or oxygen radicals
Occur during replication as a result of errors in base pairing
Point mutations
Result from base-pair substitutions in the DNA or in the insertion/deletion of a base pair
Can lead to single amino acid changes, larger changes or no change at all
Molecular Basis of Mutation (2)
Possible Effects of Base-Pair Substitution
Missense mutation
Amino acid changed
Polypeptide altered
Nonsense mutation
Codon becomes a stop codon
Polypeptide is incomplete
Silent mutation
No affect in amino acid sequence, although there is change in the genotype.
Molecular Basis of Mutation (3)
Shifts in the reading frame of mRNA
Deletions and insertions cause more dramatic changes in DNA
Lead to frameshift mutations, often leading to complete loss of gene function
Reversal of mutations
Some mutations are notoriously unstable and the mutants quickly revert back to the wild-type
Can be due to a back mutation:
Reverts back to the original base sequence
Can be due to a suppressor mutation:
A second mutation arises in the genome restoring the phenotype to that exhibited by the WT
Genetic recombination
The physical exchange of DNA between genetic elements
Homologous recombination:
Process of genetic exchange between homologous DNA from two different sources
Occurs during meiosis in sexually reproducing organisms.
Involves the exchange of genetic material between homologous chromosomes (chromosomes of the same type, one inherited from each parent).
Results in gametes (sperm or egg cells) with unique combinations of alleles.
DNA sequences have the same or nearly the same sequence allowing base pairing
Genetic Exchange in Prokaryotes
In prokaryotes, genetic recombination is observed because fragments of homologous DNA from a donor chromosome or plasmid are transferred to a recipient cell by one of three processes:
Transformation - picks up plasmids or chromosomal DNA
Transduction - tends to be chromosomal DNA
Conjugation - tends to be a plasmid
It is after this transfer, when the DNA fragment from the host is in the recipient cell, that homologous recombination may occur
Transformation
Short fragments of linear DNA bind to cell wall of competent cells
The bound DNA enters the cell and is attached to small proteins which prevent degradation
ssDNA incorporates into homologous regions of cell’s chromosome by recombination using RecA
Transduction
Transfer of donor DNA from one cell to another is mediated by a bacteriophage
Two modes:
1. Generalised transduction
DNA derived from any part of the host genome becomes part of the DNA of a mature virus particle in place of virus genome.
2. Specialised transduction
DNA from a specific region of the host genome is integrated directly into virus genome usually replacing some of the virus genes.
Generalised Transduction
Occurs during lytic cycle of phage replication
During production of new copies of phage DNA, the bacterial chromosome degrades into fragments and can be taken up by new particles during assembly
When these transducing phages infect new bacterial cells, they inject bacterial DNA which can recombine with the host chromosome
Specialised Transduction
Involves the lysogenic cycle of phage reproduction
Prophage integrates into bacterial chromosome, and during reversion to lytic cycle prophage separates from bacterial chromosome but takes some adjacent bacterial genes with it
The new phages assemble with mix of viral and bacterial DNA, which is transferred to a new host
New host survives because mixed DNA cannot be replicated, but can recombine with new host chromosome
Conjugation
Transfer of genetic material between bacterial cells requiring cell-to-cell contact
In Gram-negative cells, union is via a sex pilus
In Gram-positive cells, cells aggregate together by the recipients producing pheromones and the donors producing adhesion proteins
Important in plasmid transfer
Definitions
antimicrobials
antibiotics
antiseptic
disinfectant
preservative
selective toxicity
bacteriostatic
bactericidal
AB susceptibility
Susceptibility of individual m/o to individual antimicrobial agents varies significantly
e.g. Differences in Gram positives and Gram negatives to the penicillins
Certain broad spectrum antibiotics, such as tetracycline, are effective against both groups
Wider medical use than narrow spectrum antimicrobial
Antibiotics with limited spectrum however, may have valuable activity
Measuring susceptibility
Minimum inhibitory concentration (MIC)
Lowest concentration of antibiotic / antimicrobial needed to inhibit growth of an organism
Minimum bactericidal concentration (MBC)
Lowest concentration of antibiotic / antimicrobial needed to kill an organism
This may be equal to or higher than MIC
Principles of antimicrobial therapy
Clinical diagnosis
Signs and symptoms
Microbiological diagnosis
Microscopic, cultural, serology, molecular biology
Sensitivity tests
Provides a guide to choice of agent
Laboratory control of treatment
Factors affecting agent choice
Known or predicted in vitro sensitivity of the m/o
Patient factors
Urgency for correct therapy
Bactericidal or bacteriostatic action
Drug pharmacokinetics
Combinations
Cost
Combination therapy
Advantages | Disadvantages |
Achieve a broad and bactericidal spectrum in severe and undiagnosed infection | Increased likelihood of side-effects and toxicity |
Treat pathogens at multiple sites in the body | Superinfection and hospital cross-infection by antibiotic-resistant strains |
Achieve synergy | Possible antagonism between agents |
Prevent resistance emerging | Increased cost |
Treat mixed infections | |
Prevent fungal overgrowth |
Mode of actions
Antimicrobials/antibiotics usually act in one of five major ways:
1.Inhibition of cell wall synthesis
2.Inhibition of protein synthesis
3.Inhibition of nucleic acid synthesis
4.Injury to the plasma membrane
5.Inhibition of essential metabolite synthesis
Inhibition of cell wall synthesis
Beta-lactam antibiotics
Penicillin
Penicillin G from P. chrysogenum (1929 – Flemming)
Florey (1939) – large scale production
Active primarily against Gram positives
Modification of structure led to semi-synthetic antibiotics (some with broader spectrums of activity)
Cephalosporins
Produced by fungus Cephalosporium sp.
Semisynthetic cephalosporins have broader activity than penicillins
Beta-lactam antibiotics – mode of action:
Beta-lactams (penicillin G, ampicillin, cephalosporins & carbapenems)
Structure based on b-lactam ring
Act by binding to enzymes involved in peptidoglycan synthesis: penicillin binding proteins (PBPs)
Inhibition of cell wall synthesis leads to death by cell lysis or by a non-lytic mechanism
Glycopeptides
Vancomycin
Interferes with cross-linking of peptidoglycan preventing cell wall synthesis
Often used as a last line of defense against MRSA
Isoniazid
Important growth factor analog with a very narrow spectrum of activity
Effective only against Mycobacteria as interferes with synthesis of mycolic acid
Inhibition of protein synthesis
Antimicrobials act by interacting with the ribosome and disrupting translation
Specific and may involve binding to rRNA
Many antibiotics specifically inhibit ribosomes of m/o from one phylogenetic domain
Aminoglycosides
Bactericidal drugs (including streptomycin & gentamicin)
Act by binding to 30S subunit of ribosome
Distorting its shape and preventing translation of mRNA to the protein
Tetracyclines
Actively taken up by bacterial but not mammalian cells
Bacteriostatic drugs that bind to 30S subunit of ribosome halting protein synthesis
Chloramphenicol
Bacteriostatic drug that binds to 50S ribosomal subunit inhibiting peptidyl transferase activity
This prevents elongation of the polypeptide by blocking peptide bond formation
Macrolides
Bacteriostatic drugs that include erythromycin
Bind to 50S ribosomal subunit
Mode of action thought to be via inhibition of translocation
Inhibition of nucleic acid synthesis
Chloramphenicol
Bacteriostatic drug that binds to 50S ribosomal subunit inhibiting peptidyl transferase activity
This prevents elongation of the polypeptide by blocking peptide bond formation
Macrolides
Bacteriostatic drugs that include erythromycin
Bind to 50S ribosomal subunit
Mode of action thought to be via inhibition of translocation
Quinolones
Antibacterial compounds interfering with DNA Gyrase – bactericidal
Fluoroquinolones (e.g. ciprofloxacin) can treat both Gram negative and Gram positive infections
Rifamycins
Inhibit transcription by inhibiting RNA synthesis
Rifampicin binds to β-subunit of RNA polymerase
Disruption of Plasma Membrane Integrity
Polymyxins
Interact with cytoplasmic membrane leading to structural distortion, segments released and integrity destroyed
Polymyxin B, Colistin (Polymyxin E)
Often used as last resort due to side effects
Inhibition of Essential Metabolite Synthesis
Sulphonamides
Synthetic antimicrobial drugs
Competitively inhibit folic acid synthesis due to structural similarity to para-aminobenzoic acid (PABA)
Antifungal agents
Eukaryotes
Pose special problem for successful chemotherapy
Drug toxicity a problem
Can target unique fungal structures or metabolic processes
Fungal infections in immunocompromised people are an increasing problem.
Examples of antifungal agents
Category | Target | Examples | Formulaton |
Azoles | Ergosterol synthesis | Clotrimazole Fluconazole | Topical Oral |
Polyenes | Ergosterol | Amphotericin B Nystatin | Oral, IV Oral, topical |
Nucleic Acid analogues | DNA synthesis | 5-fluorocytosine | Oral |
Antiviral agents
Design of successful antivirals is difficult because:
Acellular
Agents in use are restricted
Most successful approach to controlling viral infections has been through vaccination
Many viral infections clear up spontaneously in immunocompetent patients
Examples
Category | Target | Example | Virus |
Nucleoside Analogues | Viral polymerase inhibitors Reverse transcriptase inhibitors | Acyclovir Lamivudine | Herpes, Varicella zoster HIV, hep B |
Synthetic amines | Viral uncoating blocker | Amantadine | Influenza A |
Protease Inhibitors | Viral protease inhibitor | Amprenavir Indinavir | HIV HIV |
Aciclovir – Selective Toxicity
Nucleoside analog that resembles the nucleoside guanosine
When it is phosphorylated by thymidine kinase it blocks viral DNA synthesis by inhibiting viral DNA polymerase
It is only phosphorylated in cells infected with herpes viruses because of viral induction