BIOL 325 LECTURE 14 MOLECULAR ONCOLOGY STUDY GUIDE
1/95
Earn XP
Description and Tags
Exam 4
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
96 Terms
what is molecular oncology?
interdisciplinary specialty that refers to the investigation of the chemistry of cancer and tumors at the molecular scale
what is the goal of molecular oncology?
identify genes that are involved in the development of cancer
the protein products may serve as targets for novel cancer treatments or imaging scans
this is still a fishing expedition and part of it is scanning known mutants
what are the techniques for molecular oncology? what is there a big area for? why do scientists use a range of techniques?
range from genomics, computational biology, tumor imaging, in vitro and in vivo functional models
a big area for computational biology but becomes obsolete within a year and requires to be HIGHLY trained for it
uses a range of techniques to validate the role of novel candidate genes in the development of cancer
what are driver mutations? what are passenger mutations?
driver mutation is directly responsible for the overgrowth of a cell type
passenger mutations are usually caused by the driver mutation and they’re not directly implicated in the disease state but might be down the line
still important because if a third mutation occurs, the passenger mutation becomes more hazardous for ones health
what is the ultimate goal for molecular oncology?
personalized medicine
personalized medicine is truly the only way to cure cancer because no two cancers are identical
have to get the information out of one individuals cancer and translate it to the right therapy for that individual
what do clinical labs test for?
tumor markers and genes that are predisposed to cancer
what is oncology? what are malignant tumors? what are the two types of tumors? what is leukemia and lymphoma?
the study of tumors (neoplasms)
malignant tumors are cancer
tumors are solid or hematological
leukemia = fluid-based white blood cell cancers
lymphoma = WBC cancers that form solid masses (lymph nodes)
what is metastasis?
movement of tumor cells from site of origin
why are proto-oncogene and tumor suppressor genes common targets in cancer diagnosis? name common examples of each (written response)
Proto-oncogenes is a common target in cancer diagnosis as they turn into oncogenes when it mutates. Proto-oncogenes normally promote cell division or cell survival but when a proto-oncogene mutates, it gains a function that causes abnormal cellular division and increased cell survival. Common examples of proto-oncogenes are the RAS gene and HER2. Tumor suppressor genes is also a common target for cancer diagnosis because when tumor suppressor genes are mutated, it loses it function to slow cell growth and allow for uncontrolled cell growth. Common examples of tumor suppressor genes are TP53 and PTEN.
what does tissue specific mean in molecular pathology/analytical testing? what is the problem with it?
nucleic acid and protein characteristics of tissue specific markers, such as rearranged IG, or T-cell receptors in leukemia
T-cell receptors are a normal part of the tissue system in blood
problem = hard to distinguish the tumor or cancer from the normal cell/same markers present in the normal tissue
how are tumor-specific target used in molecular pathology/analytical testing?
tumor-specific targets are more cancer-specific and will only specifically pick up just the cancer because they are not present in normal tissue
understand how LOF, GOF, and chromosomal abnormalities all play roles in cancer (written response question)
Loss-of-function mutations play a role in cancer as the gene, most commonly tumor suppressor genes, loses its normal ability to suppress uncontrolled cell growth. With this, the loss of the function results in uncontrolled cell growth and prevents apoptosis in the cells. Gain-of-function mutations play a role in cancer as the proto-oncogene that turns into an oncogene acquires a new function that enhances cell division and survivability, which is not normal in a cell. Chromosomal abnormalities play a role in cancer as structural changes like translocations, deletions, and insertions can result in a gain of function or loss of function. Chromosomal number abnormalities like aneuploidy and polysomy can lead to an imbalance in the genes, resulting in enhanced cell division.
how is cancer caused by the nonlethal genetic mutation, proto-oncogene? what are the categories for oncogenes?
when proto-oncogenes are mutated into oncogenes (~100 known) where some promote cell division and others prevent apoptosis
categories for oncogenes
growth factors
receptor tyrosine kinase (RTK) which is one of the three common classes of receptors, and is a hot spot for mutations
cytoplasmic TKs,
Ser/Thr Kinases
GTPases
transcription factors
how is cancer caused by the nonlethal genetic mutation, tumor suppressor? what are the five broad categories and example them
tumor suppressor genes lose its function to slow down cell growth resulting in abnormal cell growth
more than 1,000 known tumor suppressor genes and the most common are P53 and PTEN
the five broad categories
supposed to slow cell growth/cell cycle progression and cell proliferation (2)
supposed to fix broken DNA
cannot stimulate the cell to die by apoptosis
other crucial cellular signaling functions
how is signal transduction and cellular communication involved in cancer/oncogene?
cells will sometimes divide and should work where the growth factors interacts with the RTKs and triggers cell division through mitogen
if there’s NO growth factor for a long time, then no mitosis should happen, and the cell will go through apoptosis
if the RTK is mutated, there is no need for the growth factor anymore because the cell will go through mitosis several times without the stimulation of the growth factor
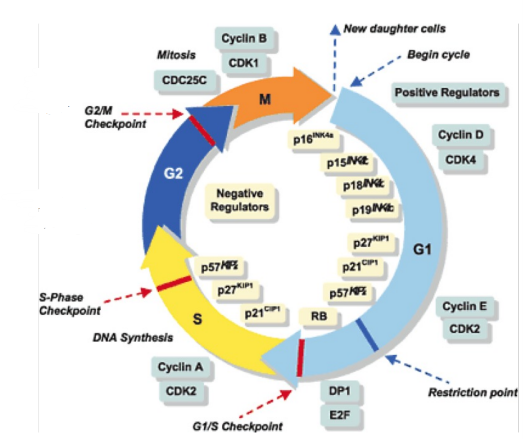
how does the cell cycle play a role in cancers related to both oncogenes and tumor suppressors? what is done if mutations are detected in the cell cycle?
the cell cycle regulates cell division and cell proliferation
cells are born in G1 and cannot exit G1 until they pass the checkpoint that sees if it is ok
involves cyclin-dependent kinases which only work if phosphorylation is present
when a cell is ready to go to the next stage, a phosphorylase and a CDK will interact and allow it to pass the checkpoint
cyclin E and CDK2 are common oncogenes and if they’re mutated, they will allow cancerous cells to pass
inside of the ring are tumor suppressors and if they’re mutated, they can no longer properly regulate the cell cycle which leads to uncontrolled cell growth
if mutations are detected, target treatment may be available or KNOWN mutations indicate high risk so affected individual screens more often
what are the three gene mutations in a gain of function for oncogenes? describe them and give an example
change in protein structure
common mutation
the proto-oncogene undergoes a point mutation, and within the gene, a protein is produced that does not degrade effectively. As a result, the gene continues to generate fresh proteins, leading to an accumulation within the gene.
ex. EGFR M277E, some lung cancers
increase in protein concentration
can have gene amplification and during replication, the polymerase puts in more copies of the same gene and each make proteins
ex. HER2/Neu breast and prostate
change in gene location
translocation, where the gene/your DNA moves to a new location under new controls/promoter
ex. t(8:14) which causes Burkitt lymphoma and t(9;22) which creates a Philadelphia chromosome that creates a new fusion protein in CML
what happens if a repair pathway is mutated in cancer?
the pathways will lose its function like mismatch repair, ss break and ds break repair
every single gene that are involved in the repair pathways are tumor suppressor mutation thats implicated with cancer
every target needs a diff therapy which is why cancer therapy is tough
what are the three targets for molecular detection of disease? describe them
antigens (antigen is on the surface of a T-cell) and mutations
disease-specific markers
translocations, point mutations, polymorphisms in tumor suppressor or oncogenes
viruses
EBV, HCV, HPV, HTVL-1
what are the 5 methods for molecular detection of disease?
hybridization, blotting
standard PCR, RT-PCR, electrophoresis
PCR with heteroduplex analysis, SSCP
real-time PCR with gene or patient-specific probes
cancer gene arrays
what is a HER2/Neu oncogene? where is it frequently amplified and what is the percent for it? what are they sensitive to? how can it be detected?
common oncogene and falls into the RTK family (intracellular kinase domain)
encodes one of family of human epidermal growth-factor receptors (hEFGR2) with tyrosine kinase activity
frequently amplified in breast and gastric cells, resulting in increased amounts of cell surface protein
20% of cancers have elevated HER2 expression
HER2-expressing tumors are sensitive to HERCEPTIN, a monoclonal antibody
herceptin can be used by any cancer with HER2 overexpression
DETECTION
HER2 protein can be detected by IHC
HER2/neu gene amplification is detected by FISH or CISH
amplification can slightly change the therapy
what does the intracellular kinase domain do? what is different about them?
phosphorylates a signal transduction pathway inside the cell
what is different about it = the outside of the cell because different growth factors stimulate all of the different external domains
how does the amplification of HER2 cause overexpression? why? what is often the cause of this?
if HER2 is amplified genetically (three copies instead of one), it leads to too much protein
normal breast cells already have HER2 protein in the membrane and is there incase an adjacent cell needs to be replaced
if there is amplification in the gene or too much protein is being made = will have EXCESS receptors in the same tissue of the cell and will trigger fast cellular division
often due to increase copy number of chromosome 17
what is the grading system for HER2 overexpression in IHC? what is the challenge when it comes to IHC?
0 = negative meaning there is NO staining
1 = negative meaning there is faint staining
2 = weakly positive meaning there is weak to moderate staining of the tumor in >10% of the cells
3 = positive meaning there is strong staining in >10% of the cells
heterogeneity is a challenge because there can be minimal staining in one area but strong staining in another on the same tissue
what are the current guidelines for IHC testing for HER2? what are the preferred samples and ideal fixative?
labs providing a testing service should carry out a minimum of 250 assays each for year for IHC detection of HER2
formalin fixed, paraffin embedded tumor tissue and buffered formalin
the use of other alternatives like bouins will prevent testing of FISH
other methods of tissue fixation can adversely affect antigen reactivity
how is FISH used to test for breast and gastric cancer? what is it looking for and what does the pink stain indicate?
FISH is used to quantitatively measure the level of HER2 gene amplification
essentially looking for DNA and uses the DAPI stain to stain the nucleus
pink staining = HER2 stain and there’s only 2 HER2 proteins per cell
what role does the EGFR family play in molecular abnormalities in solid tumors? how is HER2 apart of this?
growth receptors are often involved in cancer and if theyre constitutively activated, the cells are signaled to divide all the time
growth factors are not always activated and will only active when division is needed
HER2 is a part of this family and causes all the epithelial cells to divide
does NOT have a ligand so it works through heterodimerization with other members of the EGFR family, which leads to uncontrolled cell proliferation
more division increases the chance of mutations
driver mutations make passenger mutations
what does EGFR oncogene encode for? what are they sensitive to? how are they detected?
EGFR oncogene encodes another of the family of EGFR as HER2
common target for therapy and diagnosis
want to see which one is overexpressed to slow down its cell growth with the correct therapy
tumors with activating mutations in EGFR are sensitive to tyrosine kinase inhibitors (TKI)
DETECTION
EGFR protein = IHC
EGFR gene and chromosome abnormalities = FISH
EGFR mutations = SSCP, SSP-PCR, or direct sequencing
what is glioblastoma? what was discovered about GBM after using FISH?
glioblastoma is a highly aggressive and fast-growing brain tumor that is hard to detect → life expectancy is 2 years after diagnosis
used FISH and discovered 24% of glioblastoma showed EGFR amplification but then sequenced it and discovered it was 40-50%
also discovered that some therapies are refractory because there are other variants of EGFR
EGFR alterations are a prediction of good prognosis and will determine the overall survival of patients with IDH-wild type GBM
what is the RAS gene? what is it involved in? how is it detected?
RAS is a proto-oncogene turned into an oncogene after being mutated
RAS gene can be amplified
RAS signaling is involved in many cellular functions like growth, apoptosis, migration, and differentiation
transduction stages uses RAS
K-ras gene mutations are detected by SSCP or direct sequencing
where are the K-ras gene mutations implicated in?
implicated in various cancers like leukemia, lung, mucinous, colorectal, and pancreatic ductal carcinoma
30% of human cancers are associated with mutations in RAS genes like NRAS, KRAS, HRAS
90% of colon cancers have mutations in RAS
if patient has two particular mutations (KRAS) then the medication Cetuximab wont work
one mutation in GTP-BD results in permanent activation of RAS and cannot be deactivated
why was RAS considered undruggable until 2022?
inhibitors were not working for RAS
in 2020 the FDA granted the AMG 510, fast track designation
granted means that it was not FDA approved but approved for clinical trials only
the AMG 510 binds to KRAS-G12C RAS mutant and slowed cell growth then was approved in 2022
what are the 6 methods to diagnose oncogene activation for gene amplification or chromatin change?
FISH/CISH
WGS or NextGen
qPCR/real-time PCR
southern blot
PCR breakpoint-specific probes
break away probe fish
what are the methods to diagnose oncogene activation for mRNA/protein overabundance?
RT-PCR
RNA sequencing
northern blot
in situ staining of protein or RNAs
what might be the most productive method to scan for K-ras mutations?
next-generation sequencing (NGS)
what factors might confound its molecular diagnosis? why?
heterogeneity because you can miss a mutation
how are oncogenes activated by chromatin modification? what method do they use to detect it? what can occur with overproduction of an oncogenic product?
sometimes oncogenes activate by moving around in the genome or become exposed and expressed due to acetylation
uses FISH breakaway probes
overproduction of an oncogenic product can occur by loss of transcriptional control through chromosomal translocation
what are FISH breakaway probes? what will breakage and no breakage indicate when used?
two fluorescent probes to two targets and are very close to each other and breaks between them
can also use three different fluorescent probes to three different targets
no breakage = probes will show up next to each other
breakage = will have space between them in the nucleus
how do you detect chromosomal aberrations and molecular lesions with FISH, CISH, karyotype, or break-point probes/break away probes? can these molecular tests be used to see disease progression and treatment efficacy? (written response question)
FISH detects chromosomal aberrations and molecular lesions through the use of fluorescent probes to the targeted regions. CISH has a similar premise to FISH but uses chromogenic detection instead. Karyotyping is used to visually inspect the chromosomes by the use of Giemsa staining, which can detect translocations; however, it needs a large translocation to detect it. Karyotyping cannot detect molecular lesions. Break-away probes utilized two to three fluorescent probes to two or three target regions to determine if breakage occurred, indicating chromosomal aberrations and molecular lesions. All these molecular tests can be used to see disease progression and treatment efficacy; however, FISH is recommended for initial diagnosis.
how was FISH used to see and analyze the t(8;14) translocation? what is the t(8;14) translocation?
a green probe was designed to IGH and a red probe was designed to MYC to analyze if the translocation of chromosome 8 to chromosome 14 and vice versa occurred
if the two colors were together, it indicated a translocation and a yellow color indicated that a new hybrid chromosome was created
the translocation causes the myc oncogene on chromosome 8 to be positioned next to the IG heavy chain on chromosome 14 which causes myc to become constitutively expressed
what is the t(14;18) translocation?
reciprocal translocation between the long arms and is common in lymphomas, especially follicular
the translocation causes the BCL2 gene from B-cell leukemia and lymphoma leading it to become dysregulated and overexpressed when it comes from chromo 18 to chromo 14
how was the t(14;18) translocation detected by PCR? how is it seen on a gel?
PCR will use a downstream primer to the IG region and an upstream primer to the BCL2 gene
one primer to one chromosome and the other primer to a different chromosome
in a healthy chromosome, there will be NO PCR product
translocation in the major breakpoint region = PCR product further up the gel
translocation in the minor cluster region = PCR product further down the gel
often need multiple PCR rounds
what are chimeric genes and how might one be identified diagnostically? (written response question)
Chimeric genes are formed when two portions of chromosomes fuse together which are primarily through translocations. Chimeric genes often create a new protein that is not seen in normal tissues. An example of this would be the BCR-ABL gene that is formed through the reciprocal translocation t(9;22). Chimeric genes are identified through FISH for initial diagnosis and through PCR like qPCR and RT-PCR.
what is the philadelphia chromosome? what does the fusion protein activate and result in? why is it a good target?
a reciprocal translocation of chromosomes 14 and chromosome 18 which forms a new fusion protein called BCR-ABL
90% of patients with CML have this chromosome
BCR-ABL gene activates so many different cellular communication pathways and will lead to increased cell growth, proliferation, and increased gene transcription
results in both aberrant activity and subcellular location of the ABL protein TK, leading to cell transformation
BCR-ABL is a good target since it is not in normal cells which is the reason why CML has a great cure rate
how are translocations in hematological tumors detected?
detected with higher sensitivty using PCR
qPCR can be used to quantify the tumor load during patient monitoring
FISH is used for initial diagnosis but PCR is used for monitoring
how do you detect chromosomal aberrations and molecular lesions detected with PCR, RT-PCR, qPCR, and SSP-PCR? can these molecular tests be used to see disease progress and treatment efficacy? (written response question)
PCR is used to amplify specific DNA sequences to detect chromosomal aberrations and molecular lesions. RT-PCR turns the specific mRNA sequences into cDNA to detect any translocations, gene fusions, changes in gene expression, and mutations. qPCR detects chromosomal aberrations and molecular lesions by quantifying the miRNAs, oncogenes, telomerase, DNA, RNA and more which can also be applied to RT-PCR. SSP-PCR uses sequence specific primers to targeted regions of the molecular lesions and chromosomal aberrations at the 3’ end. These molecular tests can be used to see disease progression and determine the efficacy of treatment.
what do you do in order to get a linear trend in qPCR to ensure that the assay is working? what is it also responsible for?
take patients tumor cells and spike it with normal tissue (10x more, 100x more, 1000x more) which creates the linear trend line
also responsible for RNA isolation because as you’re adding normal cells to tumor cells, you’re also extracting RNA for RT-qPCR
essentially adding cells to cells
in order to get the standard curve for RT-qPCR, what needs to be done?
use a standard curve of transcripts of known copy numbers diluted into normal RNA
spike the RNA that was isolated from qPCr with normal RNA to create the standard curve
what happens when a tumor suppressor gene gets mutated? what may be significant in the development of human cancers? what kind of mutation are they, recessive or dominant?
tumor suppressor gene loses its function to suppress abnormal cell growth and results in uncontrolled cell growth
LOF of these genes may be more significant
recessive mutation since both alleles that code for a particular protein need to be affected (two hit hypothesis)
changes the protein structure
what is TP53/what does it encode? what does it do in the body? what does the mutated form of p53 cause?
TP53 has a normal function to assess DNA damage
encodes a “caretaker" gene” which is involved in DNA repair, induce apoptosis, transcription control, and regulating the cell cycle
binds to a double-stranded break when UV light damages DNA in the genome, which causes cell arrest and gives the cell time to repair the break
for the mutated form of p53, the signal transduction cascade cannot activate p53 meaning p53 does not interact with the dsDNA break → hopefully leads to the cells death but in other cases it causes uncontrolled cell growth
how is TP53 specifically connected to cancer? what does its indication mean? how are they detected?
2 million cancer cases each year and about 50% of them are related to the p53 mutation
can be dominant negative mutation or LOH
TP53 mutations are an indicator of a poor prognosis
DETECTION
mutated p53 protein is detected by IHC by its persistence
TP53 gene mutations are sequenced
SSCP is used to look for alternated protein confirmations and silent polymorphisms
what is inside of the p53 gene that was created? what is needed to determine the prognosis of p53 and why?
has a ton of subunits that interact with other subunits in different situations and also has a lot of different mutations
DNA and protein results often conflict, thus p53 status to determine disease prognosis required 2+ different test types
reason through some broad diagnostic differences when identifying solid tumors vs hematological malignancies (written response question)
A broad diagnostic difference in solid tumors vs hematological cancers is that solid tumors are more generally going to be caused by a mutation in the EGFR, like HER2. In contrast, hematological cancers are commonly due to chromosomal aberrations, more specifically, translocations. Hematological cancers are also more fluid-based based such as blood, compared to solid tumors, where solid masses are in organs or tissues.
what is looked for in the genes of LOF tumor suppressors?
mutations
persistent or aberrant proteins
copy number variants
WES
WGS
SNP
microarray
what is looked for in the mRNA or protein of LOF tumor suppressors?
look for presence, may not be produced
look for mutation result, such as loss of DNA repair pathway
MSI
STR analysis
what is von hippel-lindau gene?
a tumor suppressor and is necessary for normal blood vessel development
the normal function is lost by mutation in hemangioblastomas, retinal angioma
blood vessel tumors form around the CNS
autosomal dominant, 95% penetrance by age 65
what are cancer susceptibility genes and why are they important clinically? (written response question)
Cancer susceptibility genes are mutations in certain genes that MAY increase the risk of some cancers. They are also called cancer predisposing gene mutations because they are usually inherited from one parent. Cancer susceptibility genes are important clinically because knowing its inheritance can help in preventing, detecting, and treating the cancer.
what is BRCA1 and BRCA2? what happens if a women inherits one of the mutations? what does variable expressivity in a patient mean? what are the preventative measures for them?
tumor suppressor genes that encode DNA repair proteins that repairs dsDNA breaks during recombination
women who inherit a mutation in one of them have a 40% chance of developing breast or ovarian cancer in their lifetime, but it’s truly a range depending on which mutation was inherited.
variable expressivity does not mean this person has a 100% chance
preventative measures = mascetomy and frequent screening
what are the frequent mutations in BRCA1 and BRCA2? how are they detected?
BRCA1 =187delAG and 5382insC
BRCA2 = 6174delT
DETECTION
direct sequencing of both genes but need to know what you’re looking for
SSCP and SSP-PCR if you have no hints
chromosome breakage tests
how was the the 185delAG of BRCA1 detected by SSP-PCR?
used a primer specific to the normal and a red primer specific to the deletion
flanks the normal primer to amplify through the region that might have the mutation (120 bp)
the red primer will amplify the same primer and is 180 bp on a gel
what is microsatellite instability? why is it linked to DNA repair failure? what is a common way to detect MSI? (written response question)
Microsatellite instability is the production of new alleles from unrepaired replication errors. Microsatellite instability is linked to DNA repair failure because it is a preliminary diagnosis of mismatch repair failure. The mismatch repair system does not properly repair replication errors in microsatellite regions and increases the number of repeats, which leads to microsatellite instability. The common way to detect microsatellite instability is through immunohistochemistry, MSI PCR, and CGE.
what does hereditary nonpolyposis colorectal carcinoma account for? what are the mutations analyzed for and where are the genes often mutated in? what is it associated with and what is looked for?
also known as lynch syndrome = accounts for about 5% of colon and endometrial cancer, plus mutations increase the risk of many other cancers
associated with mutations in genes encoding components of the mismatch repair system, most frequently MLH1 and MSH2
MLH1 is frequently hypermethylated
mutations of the genes are analyzed for LOF
MLH1 and MSH2 are often mutated in RER
looking for microsatellite instability in both genes
85-90% of HNPCC tumors have MSI
what is replication error phenotype (RER)?
slippage in replication common at 1-3 NT repeats
the stutters are normally corrected by MMR
causes the replication machinery to slip during replication and extends the repeat
what can detect instability and give its results? how are MSI’s analyzed? what does the analysis of the five loci do? what is direct sequencing used for?
MSI PCR and CGE can detect instability by comparing PCR amplicons of the microsatellite loci to normal
results for PCR in unstable loci = tumor column has more bands compared to the normal column
results for CGE in unstable loci = spreads/doesn’t stay in one area
analyzed by assessing the stability of at least five MS loci recommended by the NCI
dinucleotide or mononucleotide repeating units
analysis of five loci determines gene function
2 out of 5 means high instability
direct sequencing is used to detect the actual gene mutation
how to detect chromosomal aberrations and molecular lesions with microsatellite instability assay? can this molecular test be used to see disease progression and treatment efficacy? (written response question)
The MSI assay can detect chromosomal aberrations and molecular lesions through their microsatellite instability, which is caused by failure of the MMR system. Specifically, the MSI assay compares the microsatellite instability in tumor tissue and normal tissue. This molecular test can be used to see disease progression and treatment efficacy.
what does it mean when an individual is heterozygous? how is loss of heterogeneity linked in the cancer realm?
those who are heterozygous with a germline mutation are predisposed to cancer
LOH happens when the other allele (normal) is lost either by deletion or silenced by methylation, and the normal allele is no longer expressed, resulting in the expression of the phenotype of the mutated allele
essentially exposes the recessive mutant allele in a hemizygous state
how is loss of heterozygosity detected by STR analysis? what are the results? how do you calculate it?
shows the loss of the heterozygous STR
compares the colon cancer tissue to adjacent normal tissue
uses fluorescent primers to PCR loci associated with cancer and uses CGE or sequencing
no LOH = peak of normal and mutant allele is the same
LOH = decreased in allele ratio of 20% or more
for sequencing, sequence both top and bottom strand and compare how much mutant allele is in normal tissue and tumor tissue
calculation = peak normal allele/peak normal allele in tumor divided by peak mt allele/peak mt allele in tumor
what is oncotype Dx? what do the scores indicate for the treatments?
a gene expression assay that is commonly used for breast cancer patients
score less than 10 for instability = hormone therapy
score 11-25 for instability = randomized hormone therapy or hormone + chemotherapy
score more than 25 = chemotherapy + hormone therapy
bad prognosis and will put you on something that will kill everything
where can tumor markers be found? what are the 6 tumor markers?
found in blood, urine, or tissue affected by cancer
tumor markers
PSA
CA125
AFP
HCG
carcinoembryonic antigen (CEA)
CA 19-9
what do liquid biopsies look for? who is it more targeted for? what is it done to see?
looks for circulating-free DNA (cfDNA) in the plasma or urine OR look for circulating free RNA and circulating tumor cells (CTC)
targeted for those who finished their therapy and will go in after 6 months for detection
done to see if there is the same MSI/cancerous DNA in the body after therapy/6 months after therapy
how are the circulating tumor cell isolated? what does it end up doing? what does it tend to do? what is FACS?
uses antibody capture using antibodies to epithelial cell-specific membrane proteins like epCAM
takes the fluid based specimen and sends the cells through a capillary tube
there will be a detector with an antibody to the targeted surface proteins of the CTCs
as the cells go by one by one, if the receptor on the membrane interacts with the antibody, the machine will suck it up
ends up counting how many cancer cells vs normal cells
tend to do to see if theres any CTC and if not, you’re in remission
FACS = fluorescence activate cell sorting
why do you want to monitor cancer concentration?
to see treatment response/to see how well the therapy is working in metastatic disease
great way to keep an eye on remission
what are the four PCR variations used in oncology?
droplet PCR = type of digital PCR
able to quantify how much starting material you have by dividing the number that are positive by the total number of wells that were analyzed
TRAP/PCR-CE
measures telomerase activity for any cancer
allele-specific, non-extendable primer blocker PCR (AS-NEPB-PCR)
Δ-PCR
uses two forward primers (external and internal) and a reverse primer simultaneously to find translocations
where does traditional PCR measure? how about qPCR? what does droplet PCR do?
traditional PCR measures at the plateau/measures how much amplicon is made at the end of the PCR
qPCR measures at the exponential phase
droplet PCR (dPCR) counts individual molecules for absolute quantification
how do you better quantify the sample for dPCR? what is the process for dPCR?
emulsify into a little droplet to better quantify how much sample and how much mutated target a patient has
isolate DNA from tissue biopsy and elute it in a certain amount of sample
best way is to take 1 microliter of the diluted sample and put it into each well
after putting the sample into 1 microliter wells, can perform amplification of the wells
PCR amplify the wells separately and analyze each separately
after amplification = will end up with a 0 or 1 score
0 = no template
1 = template
what are the 5 benefits of dPCR? explain them
good for quantification
shows exactly how many copies you have and how many cells you started with and how many are mutated
high tolerance to inhibitors
during isolation sometimes there are inhibitors so you can just dilute to 1 microliter and still perform PCR
superior precision
detects very small fold changes
increased sensitivity
detect rare mutations and low abundance targets
high reproducibility
elimate amplification efficiency bias
what are the 5 applications for dPCR?
CNV
rare mutation detection
NGS library quantification
viral load detection
gene expression analysis
what are the 6 viruses that can lead to cancer? explain them
EBV = linked to hodgkins lymphoma and detected by qPCR
HBV = test for surface antigen and core antibody
treatment of virus occurs in all cancer patients if HBV+ to prevent HBV liver destruction
HCV = screens for antibodies, curing virus improves cancer outcomes
linked to liver cancer and other liver problems
HTLV = tests for T-cell lymphotropic viral antibodies
most have no symptoms, some will develop adult T-cell leukemia-lymphoma
HHV8 (herpes 8, karposi SV) = uses qPCR
antiviral drugs improve cancer treatment
Merkel cell polyomavirus (MCPV) = uses qRT-PCR to test and is associated with poor outcomes of skin (head, limb, and trunk) cancers
what is the relation of HPV in cervical cancers? how does a Pap smear detect cervical cancers? what can tests do now for cervical cancers? what do vaccines do for HPV?
HPV has been implicated in 99.7% of cervical cancer cases worldwide
Pap smears detect abnormal cervical cells
tests can look at different strains of HPV
one particular strain can put an individual at very high risk for cervical cancer and will immediately start a preventative measure when detected
there’s also at home screening that can test for diff strains too
vaccine that prevents HPV associated with cancers
vaccines can be used after virus acquisition to prevent cancer
what is hematopoesis?
in your bone marrow, there’s a stem cell called multipotential hematopoietic stem cell aka a hemocytoblast
hemocytoblast can become a myeloid progenitor and become platelets, RBCs, or WBCs
hemocytoblast can also become lymphoid progenitor and become the B and T lymphocytes
what is needed to respond to mean pathogens? what mediates antigen recognition? what rearranges similar manner?
need to make a ton of T-cell receptors to the B-cells which is how you respond to a lot of mean pathogens
two classes of receptor proteins that mediate antigen recognition are antibodies and T-cell receptors
antibody genes (IG heavy, IG light chain, VDJ regions) and T-cell receptor genes (alpha, gamma, beta, delta) rearrange in a similar manner
recombination is normal in these cells
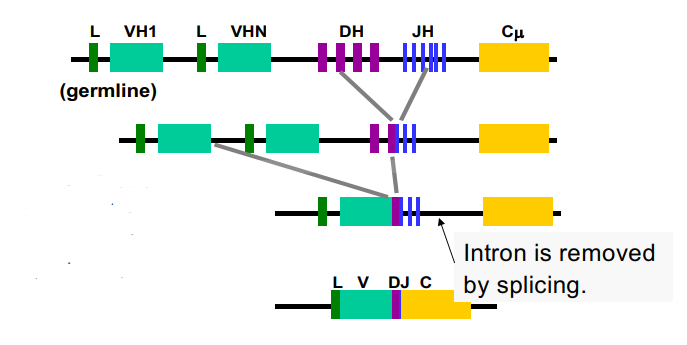
what do we have in our chromosomes related to this picture? what happens when a person is exposed to something different?
have variable regions, variable diversity joining regions, and constant regions
when a person is exposed to something different → chromosomes splice things together to make different combinations of the antibodies
by the time you get down to the mRNA, you need one of the VDJ regions
compound the diversity of the chromosome with how many different alleles exist in different populations
relates to antibody diversity
what do you have to screen for in hematological disease? what happens in most hematological diseases?
have to screen person for their genetic recombinations
what happens:
the VDJ regions go through somatic hypermutation (SMH) which is normal and occurs after the B-cell encounters the antigen
all the different cell populations you’re supposed to have, all the diff T-cell receptors, and all the different antibodies lose their polyclonality turning into a monoclonal population
what do you test for in hematological diseases? how are they detected?
tests for clonality with blood tests
normal lymphocyte populations are polyclonal with respect to IG (heavy and light) and TCR genes → all 3 may need analyzed
leukemia or lymphoma are monoclonal
depends on the clone but this is how to test/diagnose for leukemia or lymphoma
clonality is detected by protein or NA or Sblots
how are monoclonal populations detected? how are RENs used for them? what are the results?
monoclonal populations are detected by rearranged bands unique to the tumor cell population
three different RENs (EcoR1, BamH1, and Hind3) are specific to the constant regions
if there is recombination in the VDJ regions, one of their chromosomes has recombined and made a different sequence (not good) and the REN’s will cut at these different sequences at the specific REN site
essentially uses three different REN digests to show different banding patterns in the patient to show their chromosome has undergone rearrangement
EcoR1 and Hind3 results
one band in germline
two bands in rearranged
BamH1 results
one band in germline
three bands in rearranged
how was next gen sequencing used to test for clonality?
used paraffin fixed tissue and sequenced and compared the number of diff DNA seqeunces
there is a bunch of genetic info in all the cells in a healthy individual
polyclonal cell population → should be genetically different
monoclonal cell population → one particular population is overexpressed which shows patient has lymphoma or leukemia
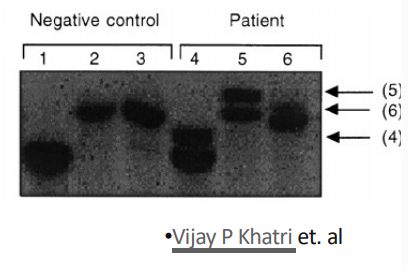
how was southern blot used to detect monoclonal lymphocyte populations? what are the advantages and disadvantages of sblot?
determined the B cell clonality in patient blood
looked at the IG chain gene with three different REN digests
negative control = different RENs should give 3 bands in total (one band for each digest)
in patient = there is a single additional (rearranged) band to indicate a monoclonal population
advantages = cheap and sensitive
disadvantage = uses radiation
how was capillary gel electrophoresis used to detect t-cell clonality? what are the results? is it expensive?
detects small sequence changes because DNA must be single stranded (denaturing)
can also use for STRs because it can detect a three NT difference (extra STR)
in a healthy control = it should look like a hill (genetically different)
y axis = abundance of the amplified fluorescent TCR-gamma (500-1500)
in a patient with monoclonal population = does not look like a hill and has multiple peaks and one peak that is overexpressed which indicates a monoclonal population
not an expensive assay and doesn’t jeopardize the health of the technician
what are the three common translocations in hematological tumors?
t(8;14)
t(9;22)
t(14;18)
what is wrong with chromosome 14?
IGH is on chromosome 14 and is a common translocation and is put next to a proto-oncogene to turn it onto an oncogene
now that oncogene is constitutively expressed
proto-oncogenes are under the control of the IGH gene, leading to uncontrolled cell growth
the translocations with chromosome 14 are commonly linked to lymphomas
how was the translocation (9;22) detected by RT-PCR? what is it looking for? what is in the gel?
designed an upstream primer to the BCR gene and a downstream primer to the ABL gene
turned mRNA into cDNA then PCR amplified the cDNA
designed a primer after ABL so it can pick up all possible translocations
looking for the expression of the fusion protein of the BCR-ABL gene
easier to pick up than mRNA
in the gel it shows all the different translocations that can occur
what is the cause of a majority of childhood cancers?
caused by fusions
80% of childhood cancers are leukemias or lymphomas and of those, more than 30% (as high as 75%) are caused by chromosomal translocations
how do you detect chromosomal aberrations and molecular lesions with RFLP and southern blot? can these molecular tests be used to see disease progression and treatment efficacy? (written response question)
Both RFLP and Southern blot can be used to detect chromosomal aberrations and molecular lesions through the use of restriction enzymes to analyze the different DNA fragments. However, Southern blotting uses radiation, and for RFLP to work, the restriction enzymes must be at the site of the mutation. Particularly, RFLP and Southern blotting are used for clonality testing and gene rearrangements in cancer patients. RFLP and Southern blot can be used to see disease progression and treatment efficacy by monitoring changes in patient DNA patterns.
how do you detect chromosomal aberrations and molecular lesions with capillary and gel electrophoresis? can these molecular tests be used to see disease progression and treatment efficacy? (written response question)
Capillary gel electrophoresis separates DNA fragments that can reveal chromosomal abnormalities like loss of heterozygosity and molecular lesions like microsatellite instability by analyzing the peaks. Gel electrophoresis can also be used to detect chromosomal aberrations and molecular lesions because it is used to resolve PCR products, which can detect deletions in chromosomes and mutations in genes. Capillary and gel electrophoresis can be used to monitor disease progression and treatment efficacy by observing the changes in the patients.
how do you detect chromosomal aberrations and molecular lesions with sequencing and microarrays? can these molecular tests be used to see disease progression and treatment efficacy? (written response questions)
Sequencing can be used to detect chromosomal aberrations and molecular lesions by analyzing the DNA sequence for mutations or any structural changes. Microarrays can detect chromosomal aberrations and molecular lesions by scanning the expression of multiple genes at the same time and comparing affected tissue to adjacent normal tissue. These tests can be used to see disease progression and treatment efficacy.
what are the three genetic aberrations that cause hematological cancers? they are also common diagnostic and treatment targets (written response question)
One genetic aberration that cause hematological cancer are chromosomal translocations. Another genetic aberration is other chromosome structural abnormalities, like deletions and insertions. Lastly, another genetic aberration that can cause hematological cancers is chromosome number abnormalities like aneuploidy and polysomy.