Lecture 5: DNA Repair Pathways
1/25
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
26 Terms
Mutations and DNA damage
Spontaneous mutations
due to natural process in cells
Ex- DNA rep errors
Inuced mutations
due to interactions of DNA with outside agent that leads to DNA damage
UC radiation
mutations lead to genetic variation that led to evolutionary changes
Transitions and transversions
Transition mutations → replace one pyrimidine base with another, or one purine base with another
Transversion mutations→ rep pyrimidine wt purine base and vs
Point mutations→ mutation that alter a single nucleotide
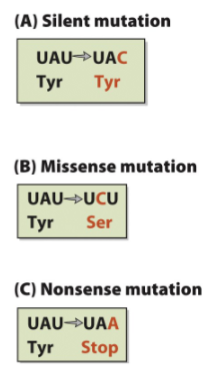
silent mutation
missense mutations
nonsense
Point mutation (sub) types
Silent mutations→ change nu without changeing AA sequence
Missense mutations→ change in protein-coding region that lead to change in AA
EX; sickle cell anemia At→ TA
Nonsense→ nu sub that leads to stop codon (premature termination of protein synthesis
-know the picture adn what final pattern looks like
Insertions or deletions can cause frameshift mutations
changes the mRNA, which leads to creation of nonfunctional protein
If it is not in pairs fo 3, it wil cause frameshift mutation
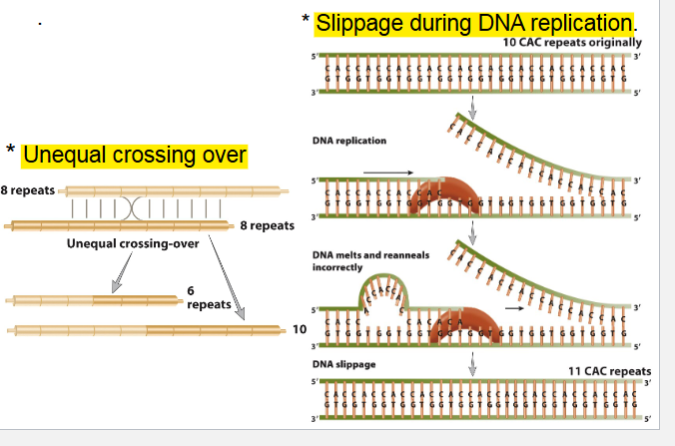
Repeat expansion found in 2 diff mechanism
Unequal crossing over
Slippage during DNA replication
3 classes of DNA damage
single base change
Deamination
structural distortions
DNA backbone damage
Single Base Change
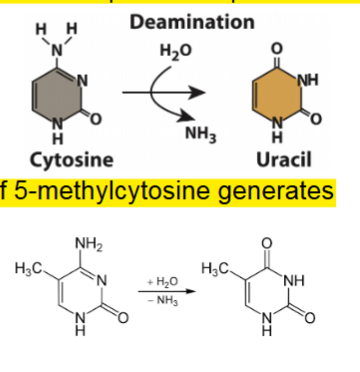
Deamination
most frequence kind of hydrolytic damage
deamination of 5-methylctosine generates thymine
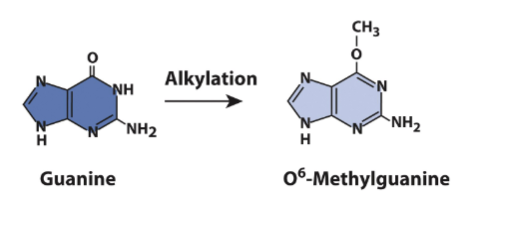
alkylation agents(ex: nitrosamines) lead to creation of O6-methylgyanosine
mispairs with thymine
GC→GT→AT point mutation after DNA Rep
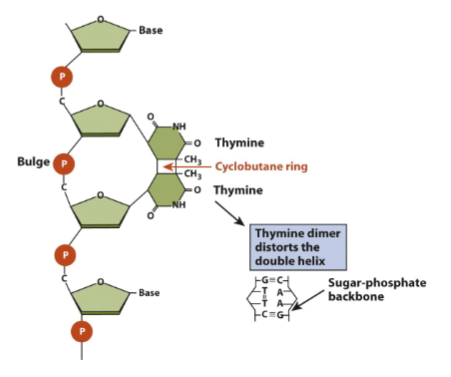
Structural distrtion
UV radiation cleates cyclobutane ring btw thymines →T-T dimer impacting double helix and block transcription and replication
DNA backbone damage
Formation of abasic sites
loss of nitrogenious bae
generates spontaneously by creation of unstable base adducts
Double-standard DNA breaks
due to ionizing radiation and a lot of chemical compounds
leads to DNA damage
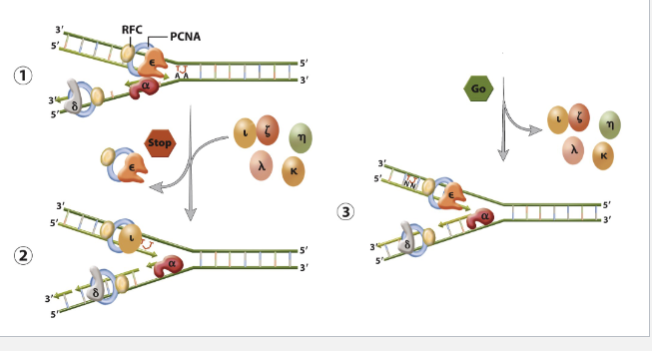
2.lesion bypass
Translesion synthesis (TLS)
error prone DNA polymerase replace replicative polymerases and copy paste with damaged DNA
DNA polymerase (n) performs translesion syn past TT dimers by inserting AA.
Direct reversal of DNA damage
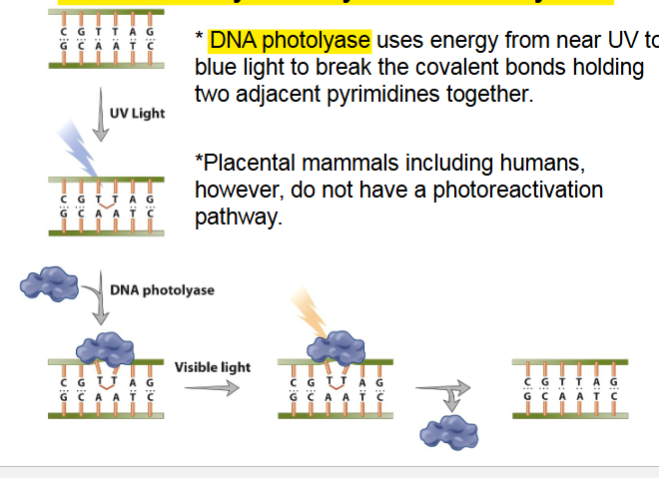
Reversal of thymine-thymine dimers by DNA
DNA photolyase use energy of blue light to break covalent bonds holding 2 pyrimidines together
humans do not have photoreactive pathway
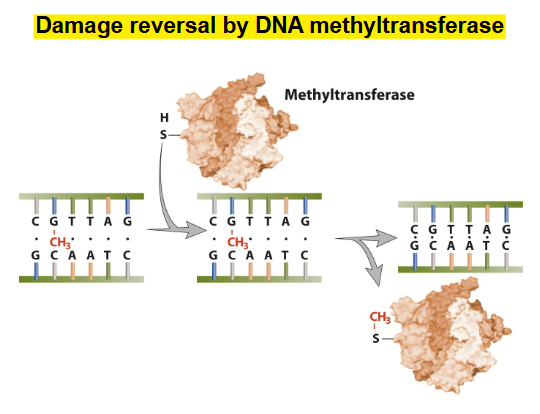
-BUT
DNA methyltransferase does damage reversal
4. Repair of single base changes and structural distortions by removal of DNA damage.
Pathways for repair of single base changes and structural distortion
single base change
base excision repair
mismatch repair
structural distortion
nucleotide excision repair
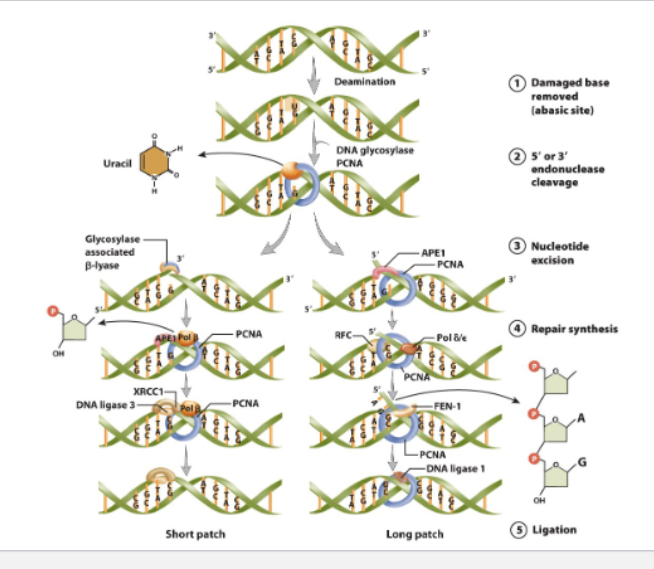
single base change
base excision repair
Mismatch repair
Base excision repair
DNA glycosylase recognizes and excises the damaged base
endonuclease Cleves the phosphodiester bond at either 3’ or 5’ of abasic site
removes 1-10 nucleotides
DNA polymerase replaces missing nucleotides
DNA ligase seals gap
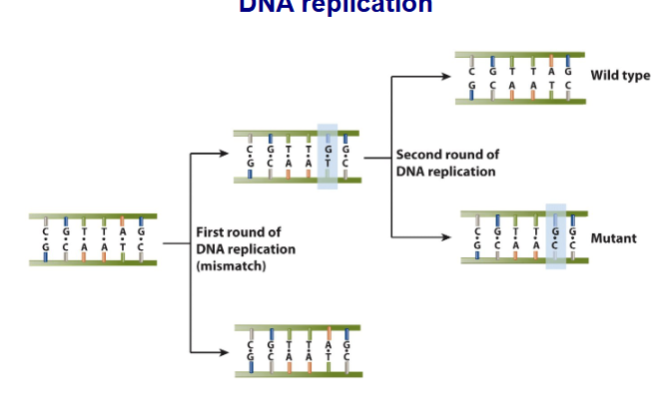
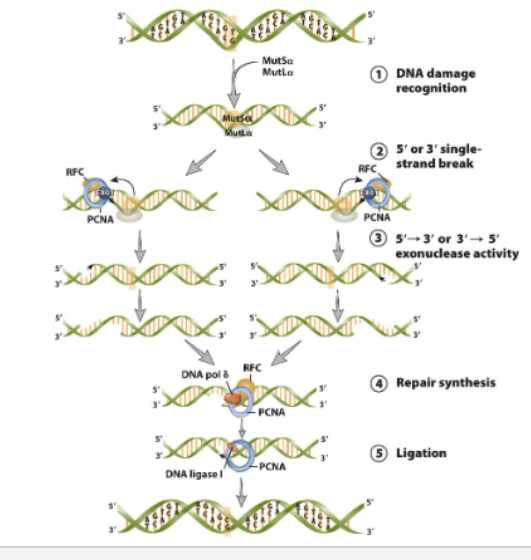
Mismatch repair pathway in mammalian cells
Mismatch error due to DNA duplication
rec by MutS”alpha”/ MitL”alpha”
Single-strand break of 5’ or 3’ generated by EXO1 with help from PCNA and RFC
5’→3’ or 3’→5’ exonuclease activity of EXO1 removes mismatch and other nucleotides
5’-3’ repair synthesis mediate by DNA polymerase
remaining gap ligased by DNA ligase 1
Recurring theme in DNA Repair
Hand-off of damaged DNA from a complex with nuclease activity to a complex with polymerase activity to a complex with ligase activity.
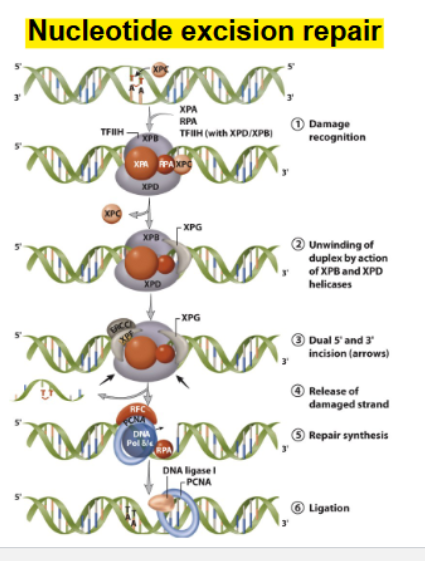
structural distortion
nucleotide excision repair
nucleotide excision repair
Repair of structural distortion
from T-T dimers from UV irradiation
GGR pathway (Global genome repair)
repair of lesions in whole genome
TCR pathway (Transcription couples repair): repair of lesions in the transcribed strand of active genes.
Xeroderma pigmentosum and related disorders: defects in nucleotide excision repair
autosomal recessive disorder
inc risk of sunlight-induces skin cancer
Defects in nucleotide excision repair or in T-T dimer translesion repair.
Double-strand break repair
due to oxygen species, ionizing radiation, and the free radical chemicals made
repaired by homologous recombination or nonhomologous end-joining
Repair by recombination
Homologous recombination
repair double strand break by getting genetic material from undamaged homologous chromosomes
Nonhomologous end-joining (NHEJ)
rejoins double-strand break via direct ligation of DNA end withNO any prior sequence homology
HOM recomb has a big role in doub-str break repair in prokaryotes and single-cell eukaryotes
double strand break primarily repaired through NHEJ in mammalian cells
homologous recombination primarily servs to repair double strand breaks at the replication fork.
Homologous recombination
many roles in eukaryotic organisms
crossing over in meiosis
transposition
mating-type switching in yeast
antigen-switching in trypanosomes
dna repair
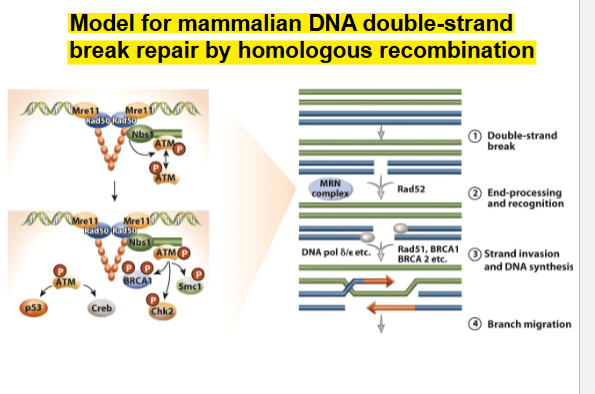
Model for mammalian DNA double-strand break repair by homologous recombination
Steps
Double-strand break (DSB) is induced by ionizing radiation.
End-processing and recognition: Recruitment of MRN (Mre11-
Rad50-Nbs1) to the DSB. The 3′, 5′ exonuclease activity of Mre11
generates 3′ ssDNA tails that are recognized by Rad52.Strand invasion and DNA synthesis. The 3′ tails invade
homologous intact sequences to generate a hybrid molecule.
Missing sequence information at the DSB is restored by DNA
synthesis.Branch migration
Holliday junction resolution and ligatio
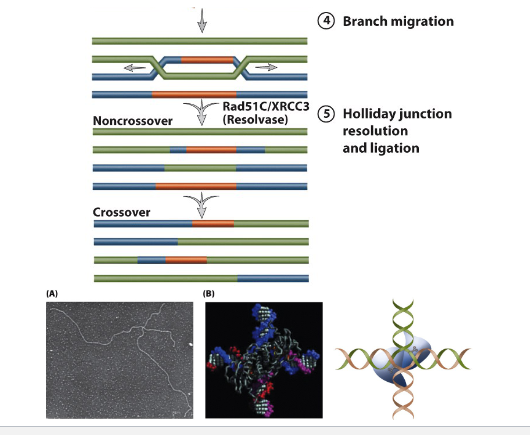
Holliday junctions
1960s→ RObin made a model of general recombination based on genetic data from fungi
2 duplexes by exzyme complex called Resolvasome
in E.Coli RuvABC resolves Holliday junctions
Rad51C required for Holliday junction processing in mammalian cells
Hereditary breast cancer syndromes: mutations in BRCA1 and BRCA2
5-10% all cases
mutation on BRCA1 and BRCA2" “tumor suppressor genes”
risk of breast (ovarian) cancer
BRCA1: 50-87%
BRCA2: 15-44%
Nonhomologous end-joining
2 broken ends is ligated together even if they came from same chromosome
double-strand break repair leads to mutations and Indels (inserts/deletions)
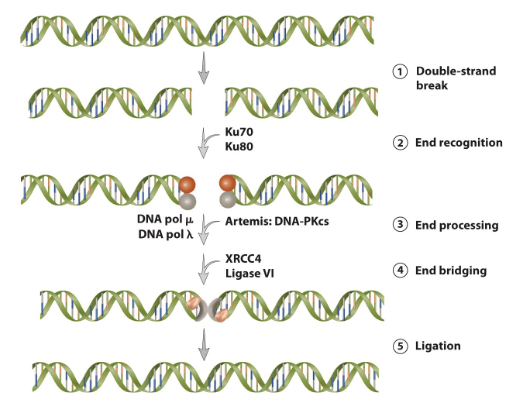
Model for mammalian DNA double-strand break repair by nonhomologous end-joining
Double-strand break.
induced by ionizing radiation.
End recognition.
Broken ends are recognized by heterodimers of
Ku70/Ku80.
End processing.
endonuclease Artemis activated by the DNA-dependent protein kinase catalytic subunit (DNA-PKCS). DNA polymerase (pol) “u” or pol “y” fill-in gaps and extend 3′ or 5′ overhangs.
End bridging
Ligase complex XRCC4-DNA ligase IV is recruited to the damaged site and forms a bridge.
Ligation:
broken ends ligated by the XRCC4-DNA ligase IV complex.