TRANSCRIPTION
1/23
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
24 Terms
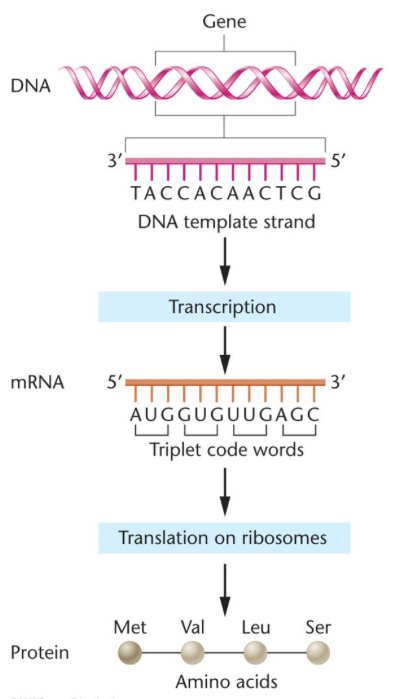
Central dogma
gene expression
Structure
indicates function
Transcription
transfer of information from DNA → RNA
RNA polymerase reads DNA template strand & uses complementary base pairing rules to synthesize RNA
Translation
cell synthesizes protein
Major steps of transcription
initiation
elongation
termination
Initation - E. coli
sigma (part of RNA polymerase holoenzyme) binds to promoter sequences
positions RNA polymerase core enzyme on the DNA template strand at the transcription start site
RNA polymerase starts synthesizing RNA in 5’ → 3’
sigma dissociates from the core enzyme after transcription starts
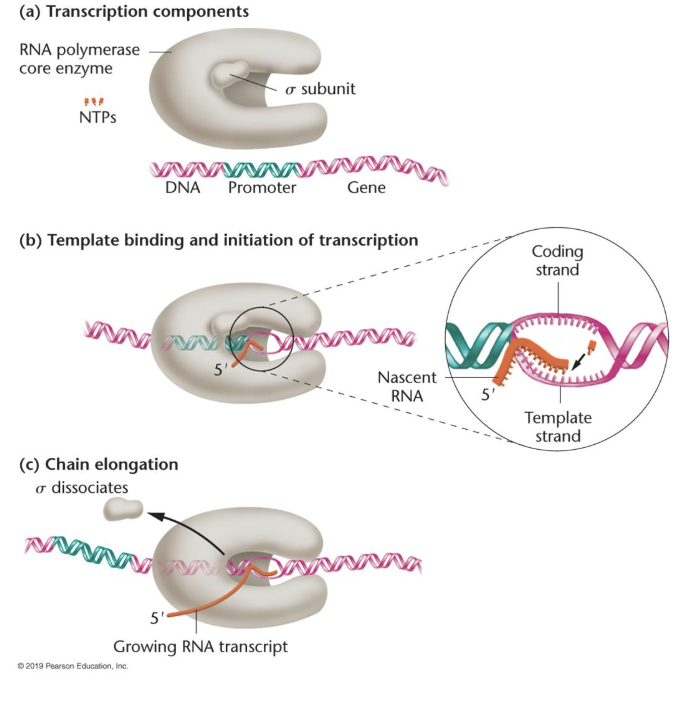
Core
RNA polymerase alone, can synthesize RNA
Holoenzyme
core + sigma factor, can recognize promotors
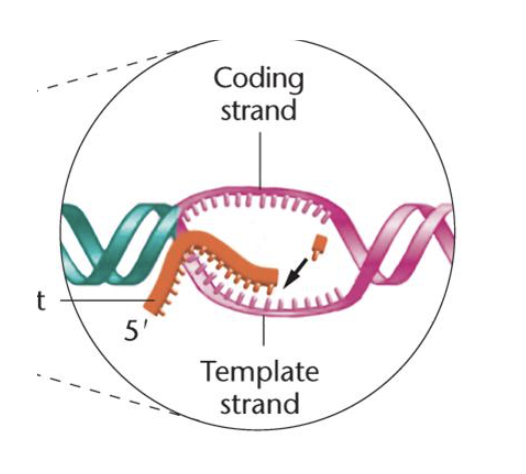
Elongation - E. coli
reaction is catalyzed by RNA polymerase core enzyme
NTPs added to the 3’ end of the nascent (new) RNA
Polycistronic
E. coli mRNA
one mRNA contains the sequence information from several genes
one mRNA encodes several proteins
allows for tight regulation of protein synthesis for proteins that are in the same pathway
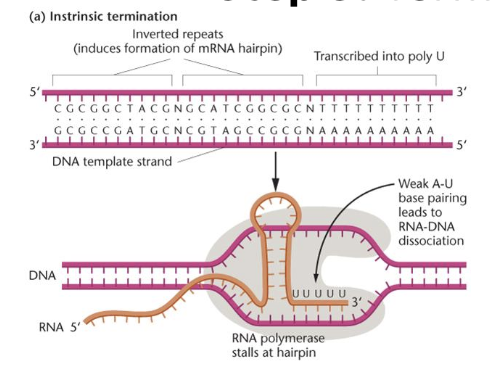
Termination - E. coli
intrinsic termination
rho-dependent termination
Intrinsic termination
hairpin structure in RNA causes RNA polymerase to stall
RNA-DNA duplex dissociates due to a weak dissociation between the RNA and DNA
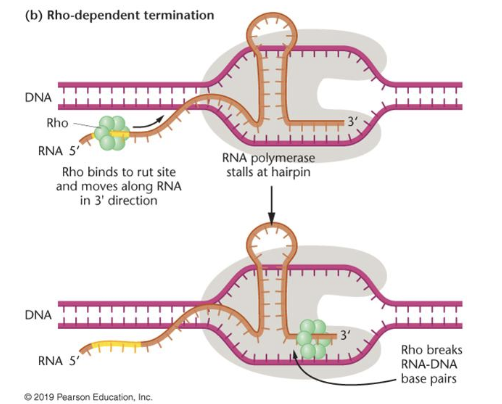
Rho-dependent termination
hairpin structure in RNA causes RNA polymerase to stall
Rho (helicase) unwinds the RNA-DNA duplex
Prokaryotic transcription
1 type of RNA polymerase
lots of copies
transcription and translation processes are near each other
polycistronic mRNA (several proteins encoded in one mRNA)
limited RNA processing
Eukaryotic transcription
3 different types (classes) of RNA polymerase (I, II, III)
transcription and translation are temporally and spatially separate by nuclear envelope
monocistronic mRNA (one protein encoded in one mRNA)
mRNA is highly processed (5’ cap, splicing, poly A tail) before being exported from the nucleus
Polycistronic
codes for multiple proteins
Monocistronic
codes for one protein
Classes of RNA polymerase in eukaryotes
RNA polymerase I - rRNA
RNA polymerase II - mRNA & snRNA
RNA polymerase III - tRNA
each are recruited to their genes by dedicated transcription factors
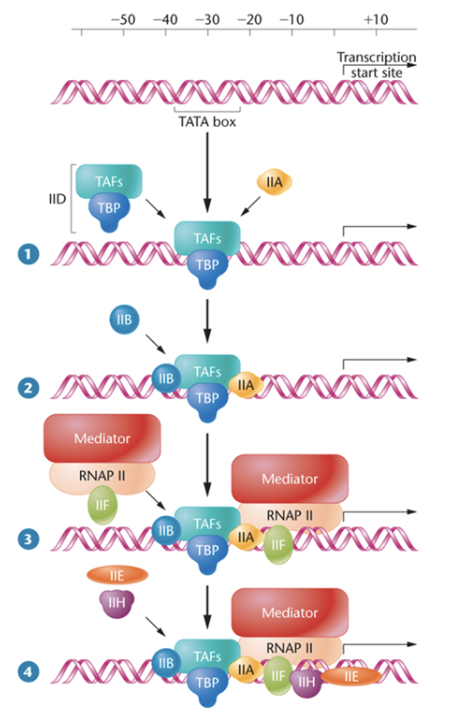
Initation - eukaryotes
general transcription factors bind to the core promoter
GTFs are named according to the polymerase with which they interact
Ex: TFIIA
GTFs recruit RNA polymerase to form the preinitiation complex (PIC)
RNAP cannot locate a gene on its own
recruitment of polymerase by GTFs is increased by activators bound to enhancer sequences (we’ll revisit this when we discuss gene regulation)
Elongation - eukaryotes
RNAP catalyzes NTP addition to the 3’ end of the nascent RNA strand (much like in prokaryotes)
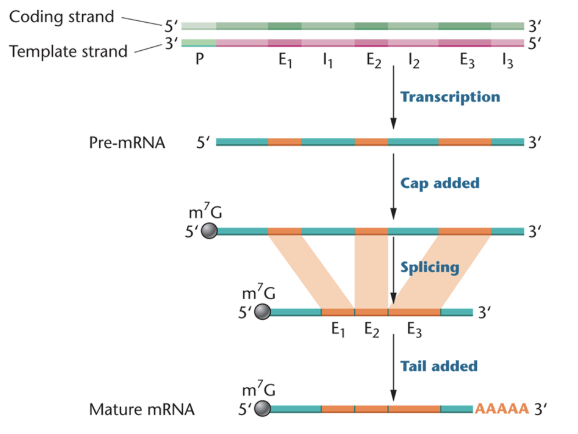
during transcription
5’ m7G cap is added to the mRNA
introns are spliced out (exons are spliced together) to form one continuous coding sequence
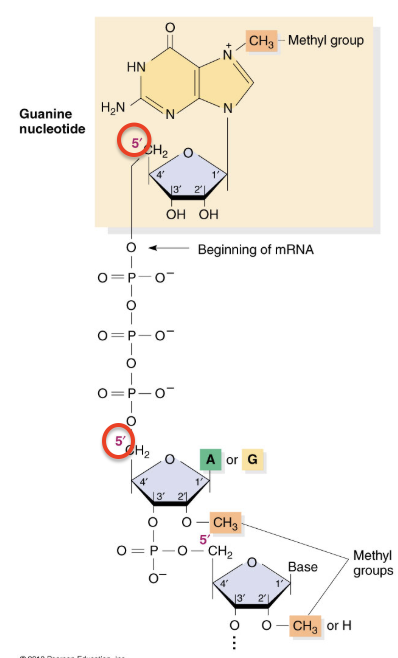
m7G cap
added to the 5’ end of the mRNA
attached to mRNA by a 5’-5’ linkage when mRNA is 20-30 nucleotides long
necessary for translation
protects RNA from 5’ to 3’ exonucleases
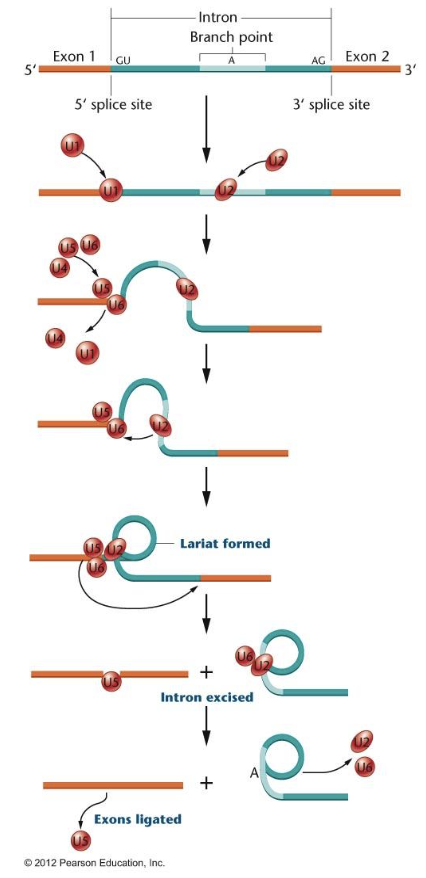
Splicing of introns
many eukaryotic genes contain long insertions of non-coding sequences called introns
introns
intervening sequence
transcribed but removed before translation
even though they are removed, introns can contain important information
exons
expressed sequence
transcribed and translated
Splicing
spliceosome is made of subunits consisting of both RNA and protein
subunits called snRNP (small nuclear ribonucleoprotein)
RNA subunits base pair with RNA sequences called splice sites within the intron
5’ splice site
3’ splice site
branch point A
loops out intron
exons covalently bonded together
introns released as a lariat and likely degraded
Termination - eukaryotes
mRNA is cleaved at the polyA cleavage site
mRNA is released from the transcription bubble
mRNA is polyadenylated
about 250 adenine nucleotides (ATPs) are added to the 3’ end of the RNA by PolyA Polymerase
called the polyA tail
PolyA tail is coated with protein
necessary for translation
protects 3’ end of mRNA from 3’ exonucleases that would otherwise degrade it