Bio Unit 2: DNA and RNA- DNA Replication, Gene Expression, Gene Regulation
1/88
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
89 Terms
Basic Nucleic Acid Structure?
Phospate group, 5 carbon sugar, Nitrogenous Base.
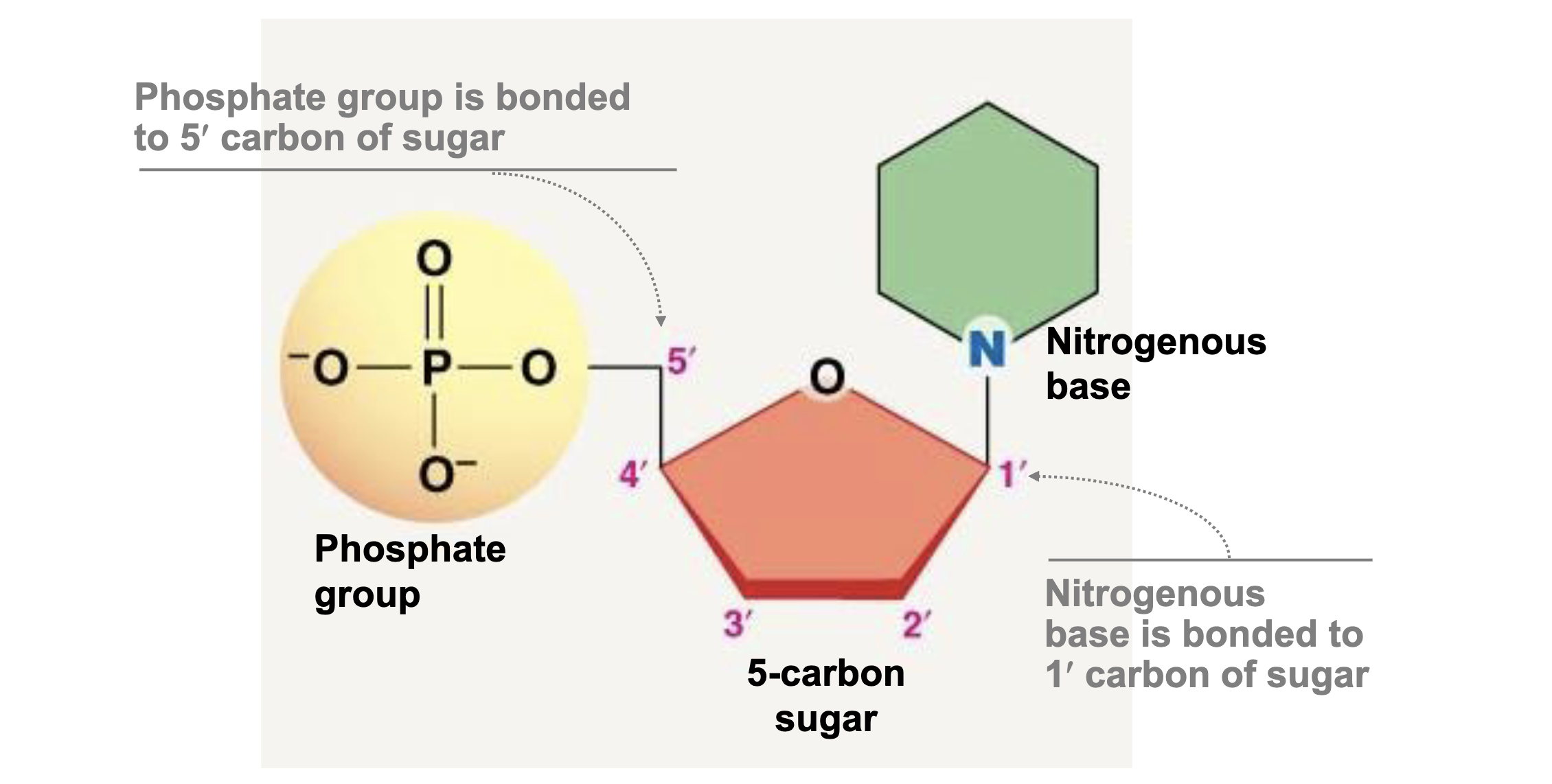
What are the main differences between DNA and RNA structure?
DNA is double stranded while RNA is single Stranded. DNA has a deoxyribose sugar (H on 2’ carbon) while RNA has a ribose sugar (OH on 2’ carbon). DNA has thymine based paired with adenine and RNA has uracil.
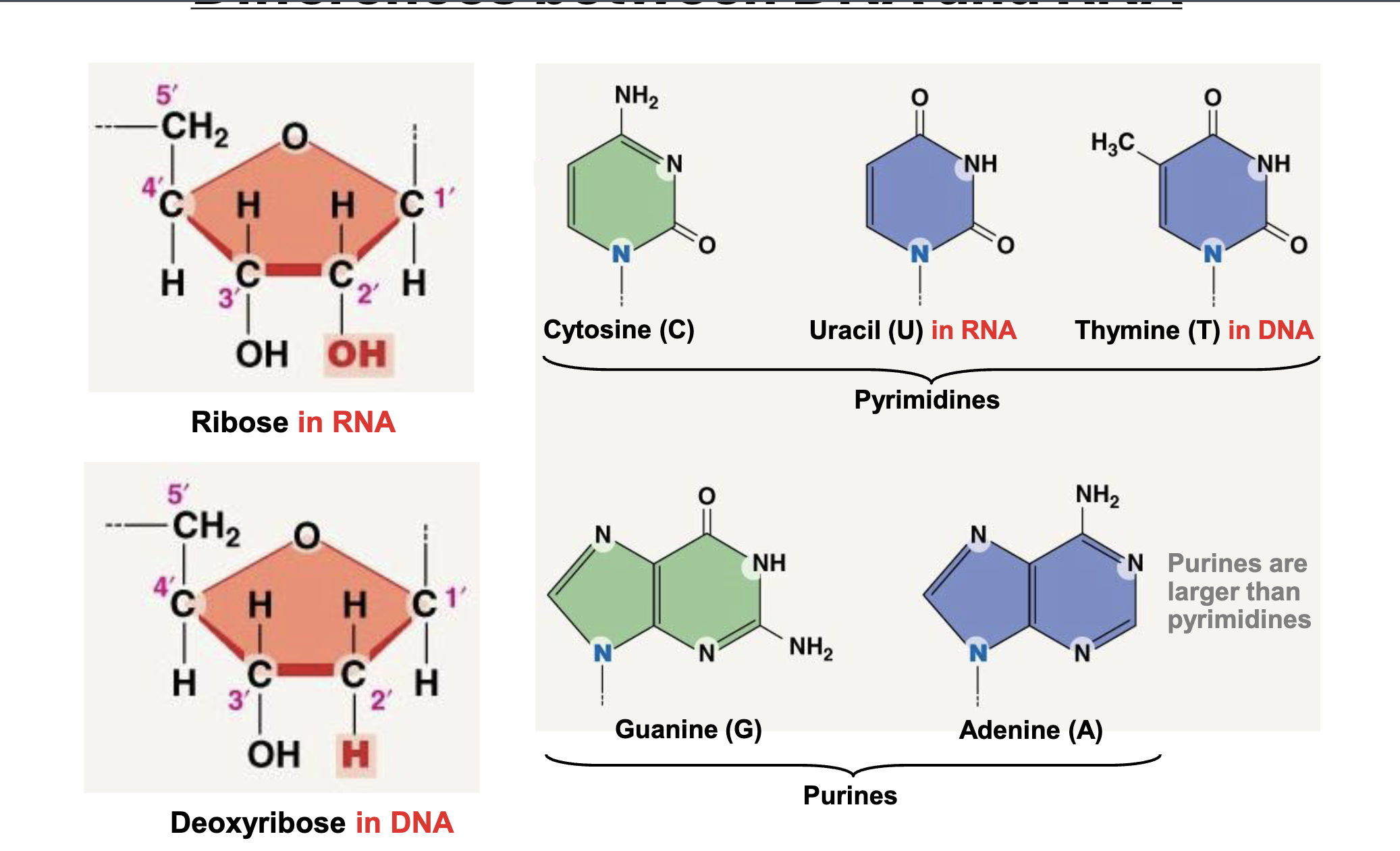
Pyrimidines
Cytosine, Uracil, Thymine
Purine
Guanine, Adenine
3’ OH role in phosphodiester bond formation
5’ phosphate bonds with 3’ hydroxyl.
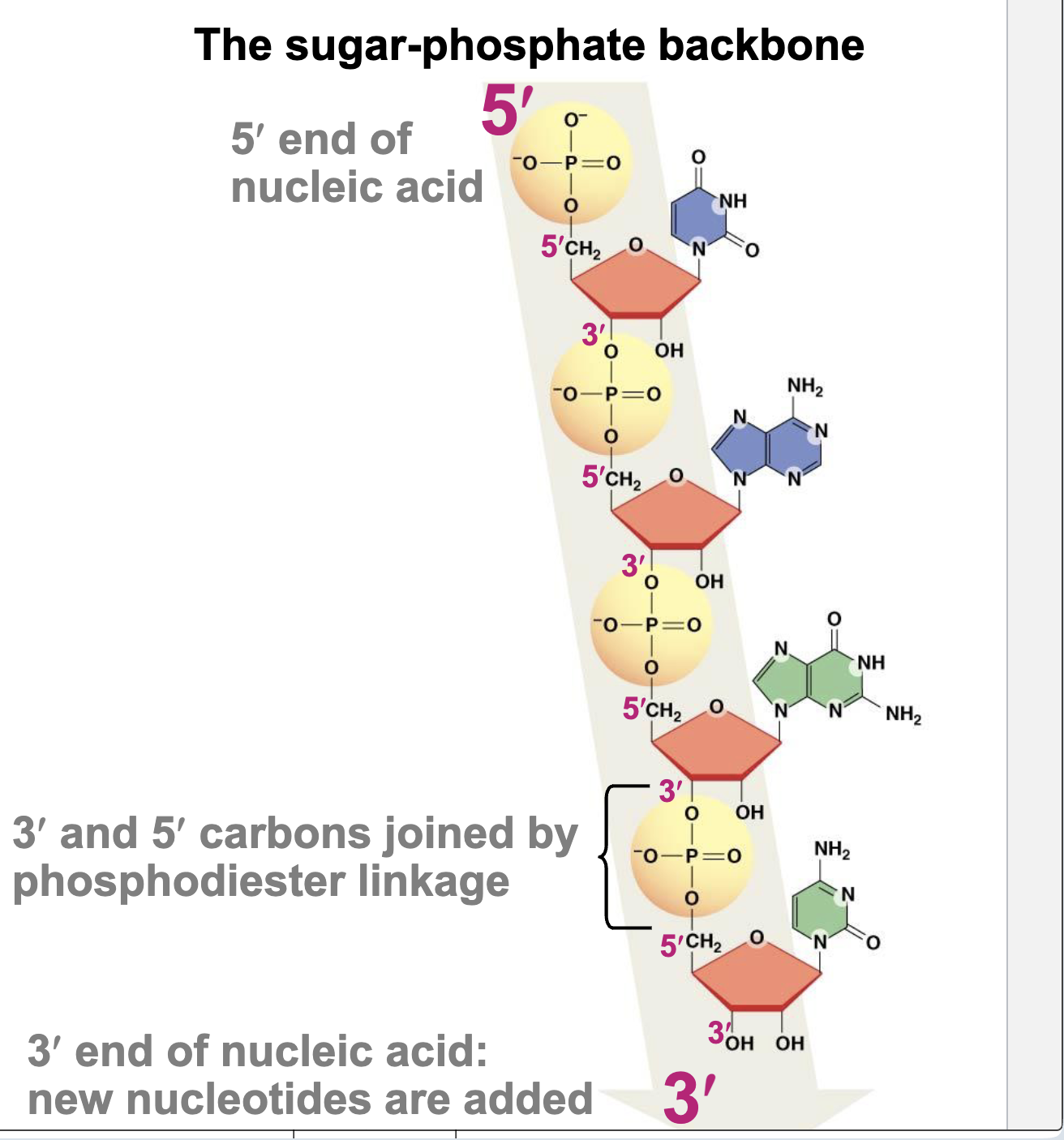
General features of nucleic acid (ends and direction)
5′ end – unlinked phosphate, 3’ end – unlinked hydroxyl, The nucleotide sequence is written in the 5′ → 3′ direction
Energetics of (5’) phosphate
The 5′ phosphate group is attached to the 5′ carbon of a nucleotide’s sugar.
Provides the reactive site for forming phosphodiester bonds with the 3′ hydroxyl of another nucleotide → builds DNA/RNA backbone.
Involved in ligation and energy transfer during replication and repair (bond formation releases pyrophosphate).
Carries negative charge, influencing molecule stability and enzyme recognition.
Energetics: ATP’s phosphoanhydride bonds release energy when hydrolyzed due to reduced charge repulsion, resonance stabilization, and hydration of products.
Bond formation: Phosphates form phosphoester and phosphodiester bonds in DNA, RNA, and membranes; phosphorylation activates molecules and regulates enzymes.
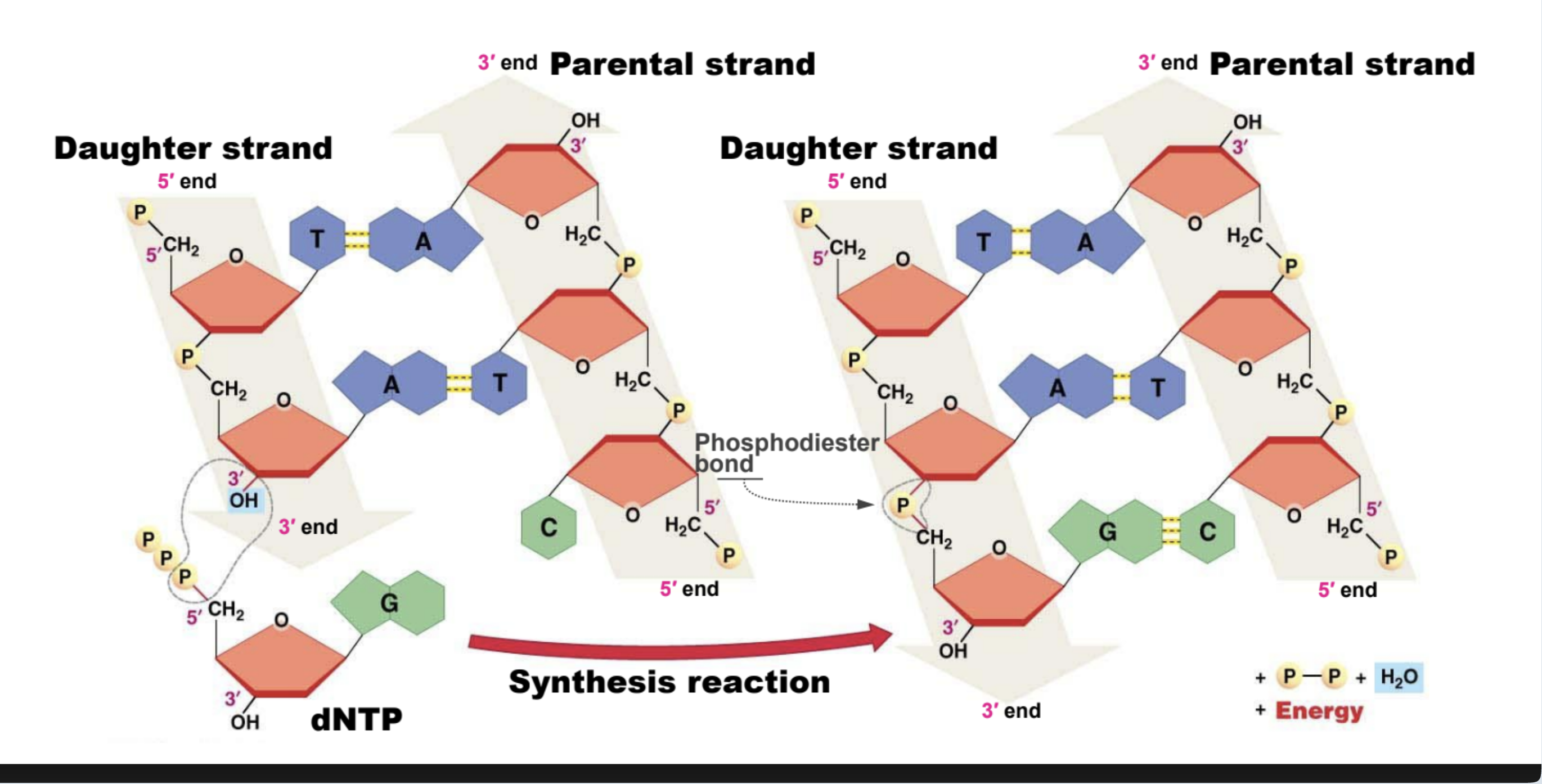
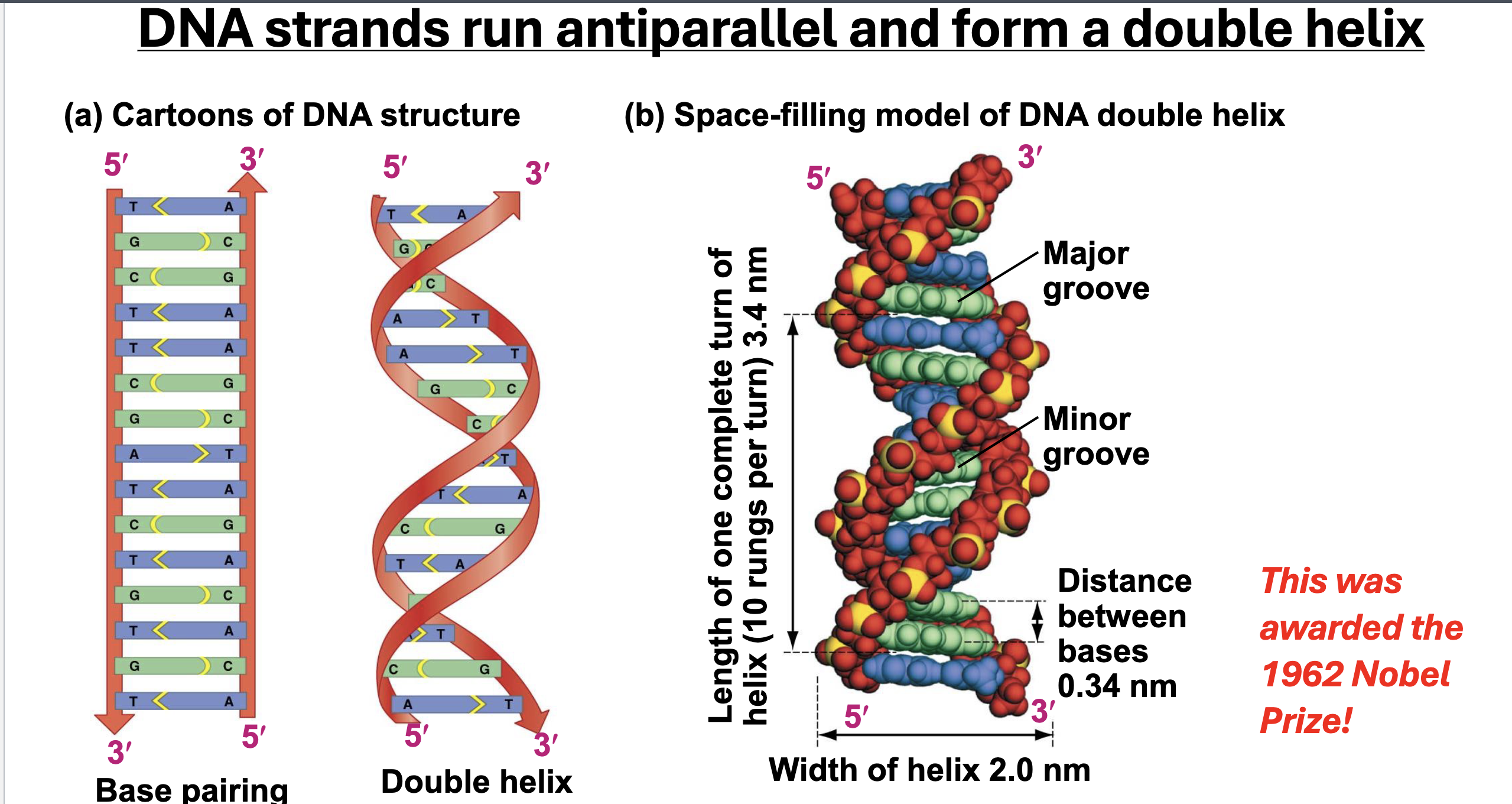
Characteristics of a DNA strand
Base Pairing ({A,T}; {G,C}), Double Helix, Directionality, Antiparallel Strands
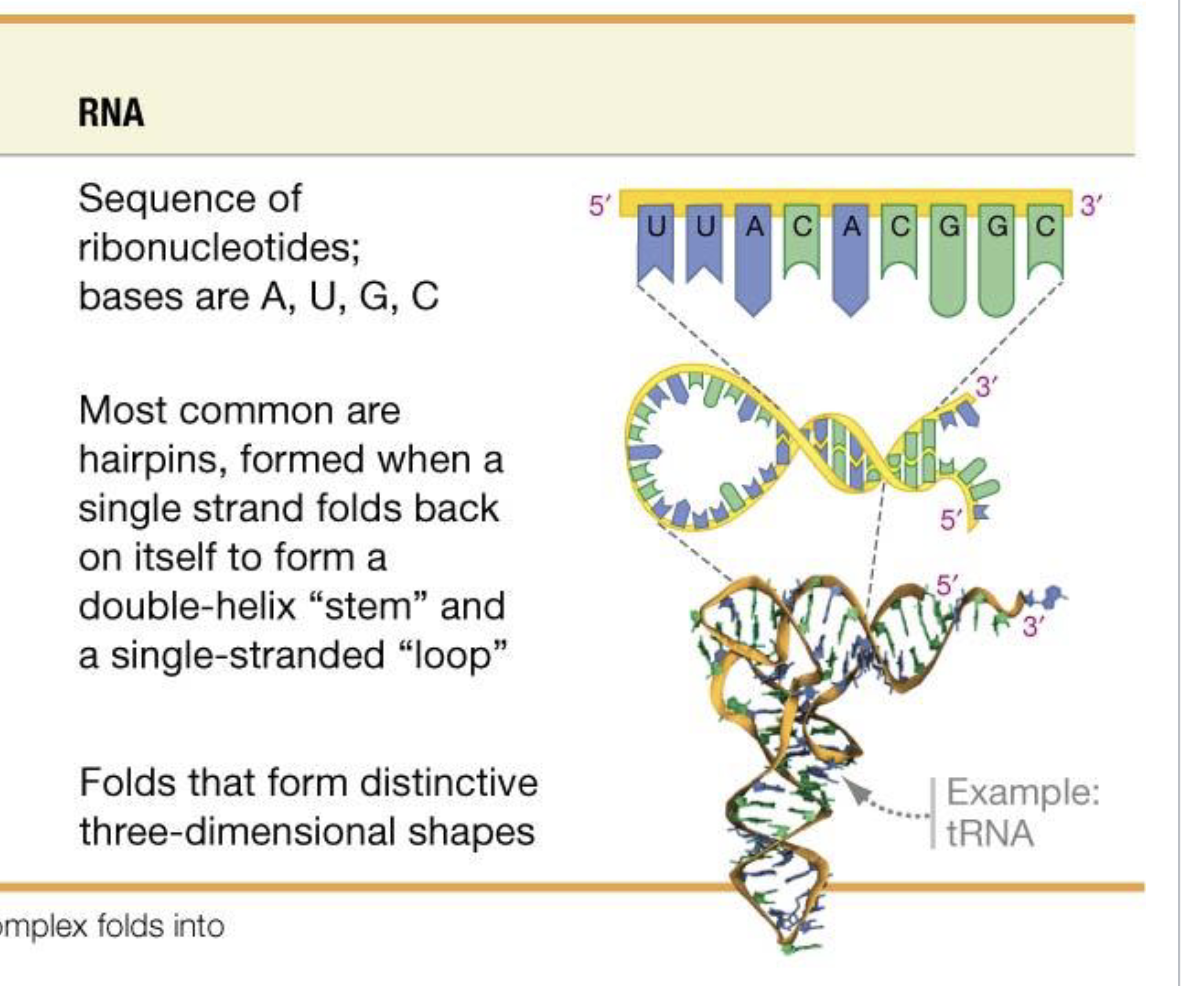
RNA Function and Characteristics
RNA is less stable and has more reactivity than DNA
RNA usually exists as a single-stranded molecule but can hydrogen bond with itself to form 3-dimensional structure
RNA can act as an information messenger, translator, enzyme and more.
Still has 5’ and 3’ directionality
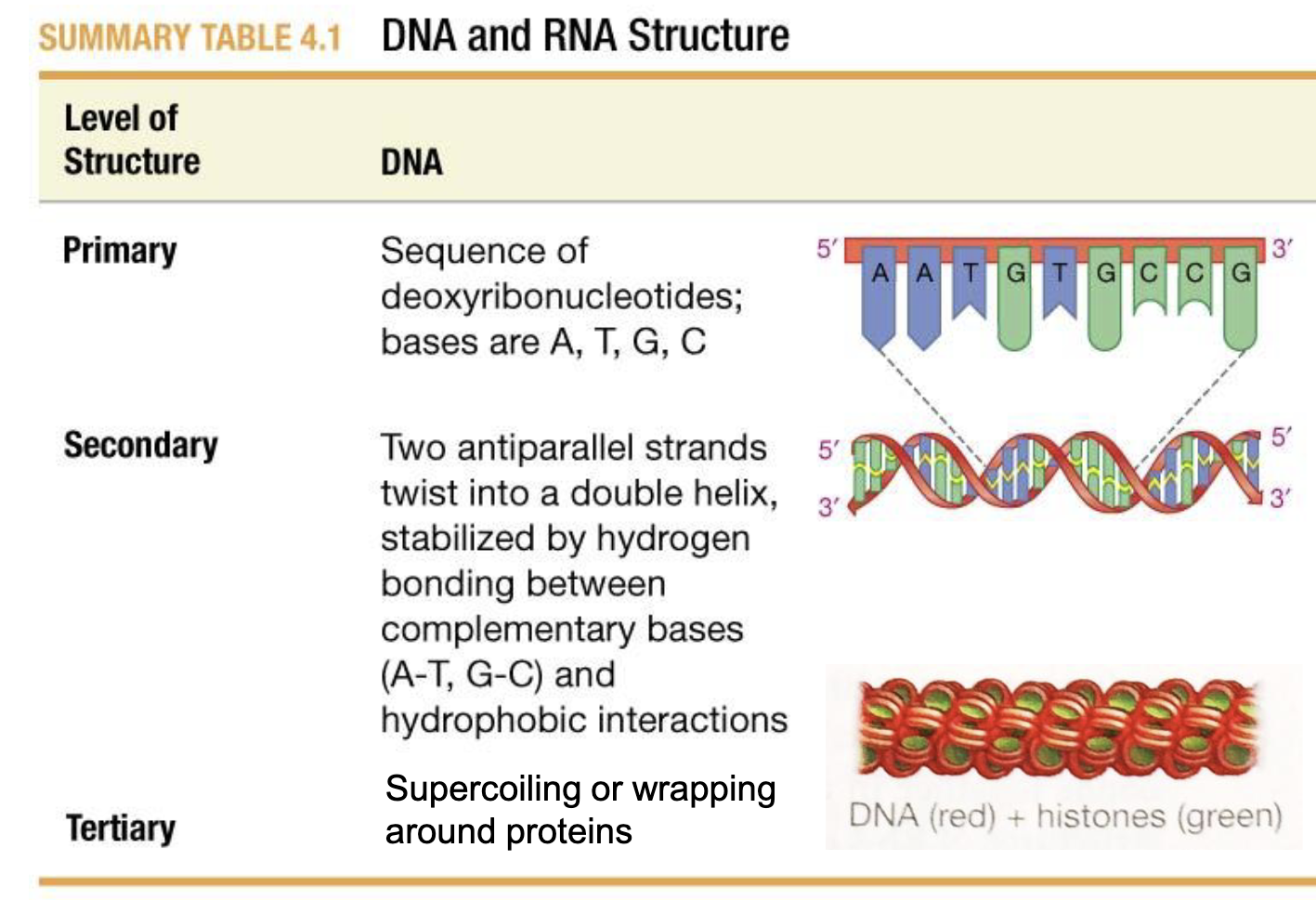
DNA Primary, Secondary, and Tertiary structure
Primary: Base Sequence
Secondary: Two antiparallel strands twisted into a double helix, stabilized by h-bonds between complimentary base pairs, and hydrophobic interactions
Tertiary: Supercoiling (Coiling on itself) or wrapping around proteins
RNA Primary, Secondary, and Tertiary Structure
Primary: Base Sequence
Secondary: Common are hairpins that form when RNA folds back on itself to form a double helix stem.
Tertiary: H-bond driven folds that form distinctive 3d Shapes.
What type of molecular interaction holds double stranded DNA together? How might we disrupt this molecular interaction?
Hydrogen Bonding. Heat (or extreme PH or Chemical Denaturant) is common to denature DNA.
Meselson–Stahl experiment
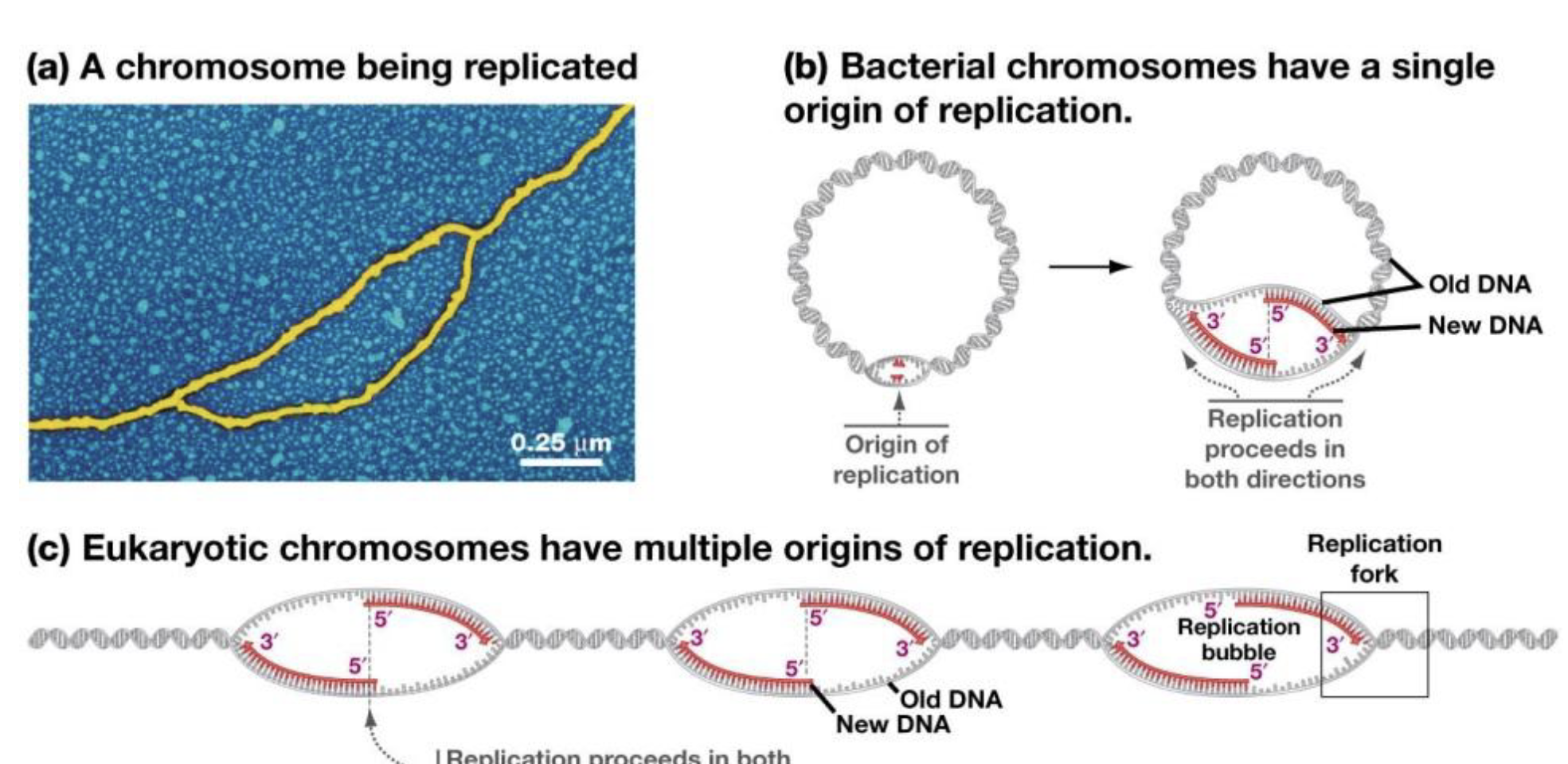
Replication Bubbles
Origins of Replication
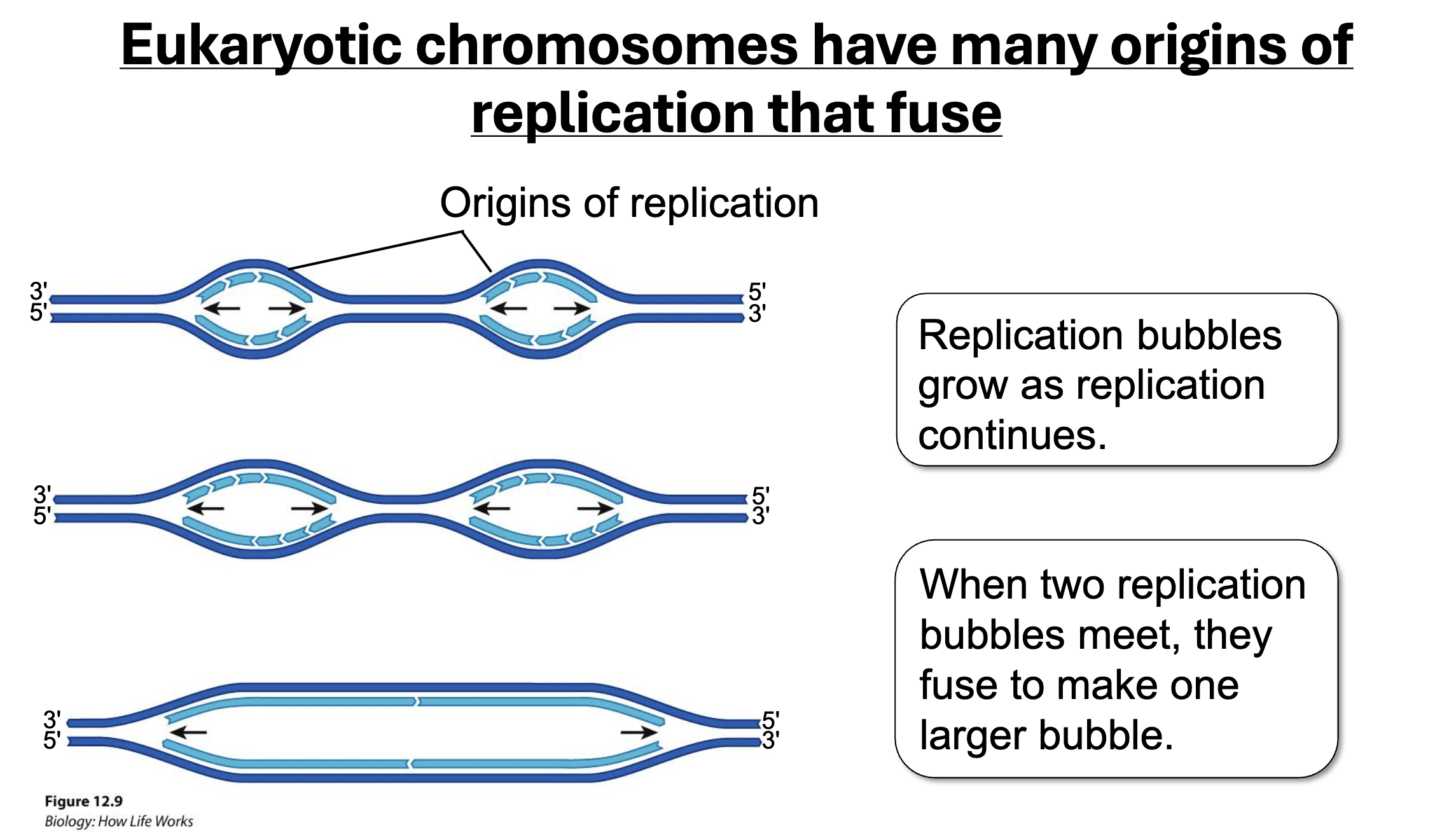
Difference between prokaryotic and eukaryotic origins of replication
Prokaryotic: Single Origin
Eukaryotic: Multiplie Origin
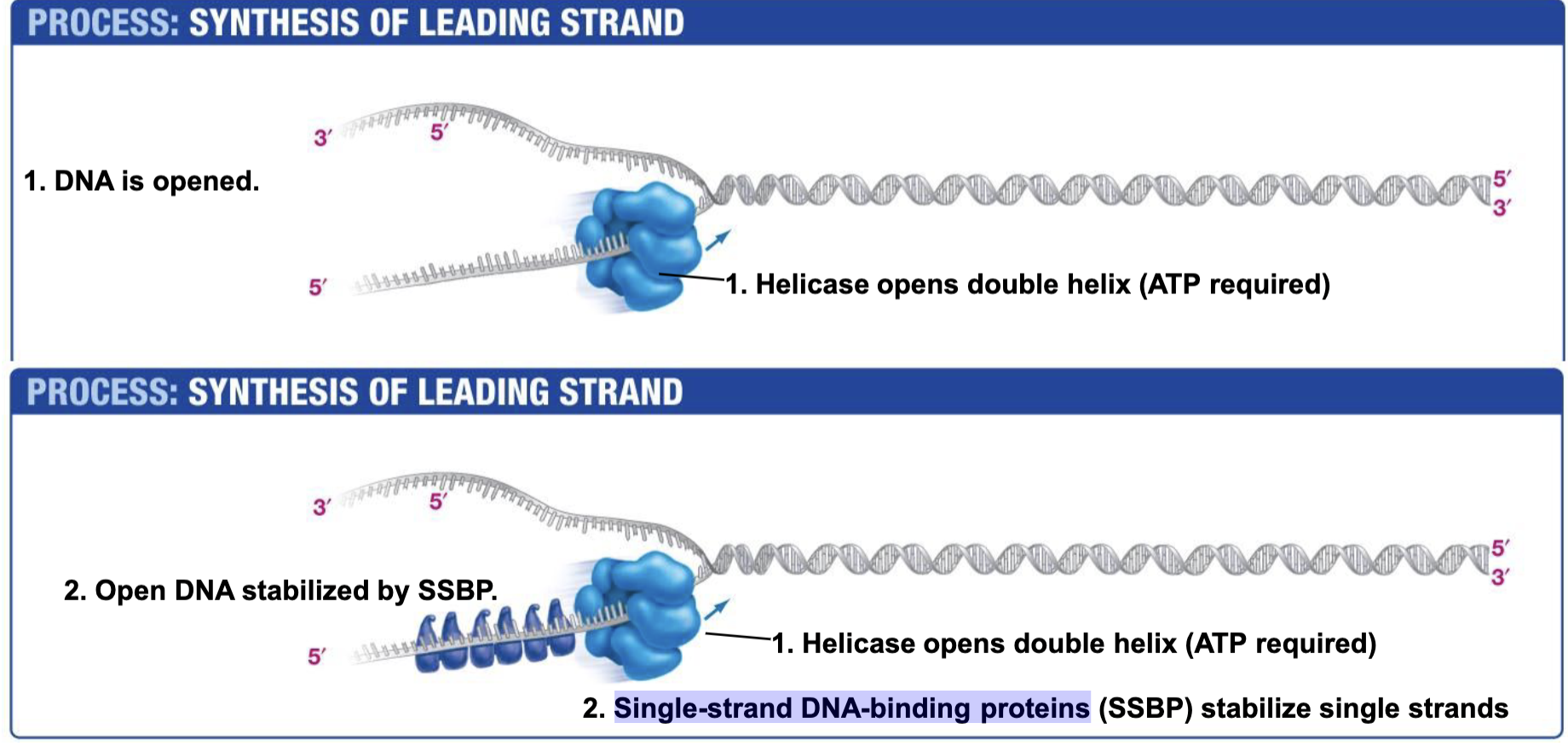
Helicase
Opens double helix; ATP NEEDED!!!! (Energy to break base pair h-bonds)
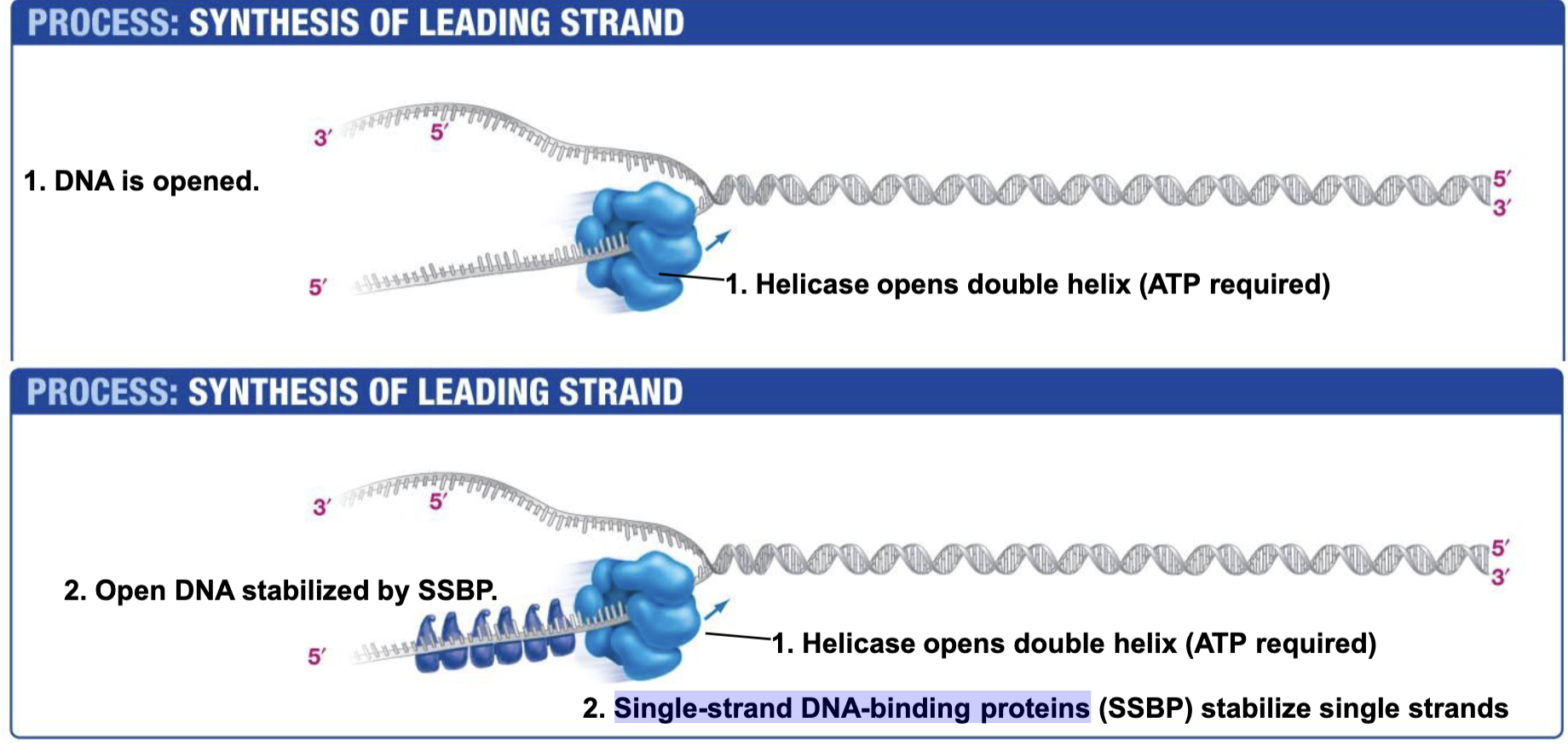
Single-strand DNA-binding proteins. SSBP
Single-strand DNA-binding proteins are needed to prevent two strands of single stranded DNA from hydrogen bonding back together once they have been unzipped by helicase.
Topoisomerase
Topoisomerase relieves the over twisting caused by helicase unzipping the DNA double Helix.
Works by breaking DNA ahead of helicase and eventually rejoining the DNA when helicase (and other replication machinery) catches up.
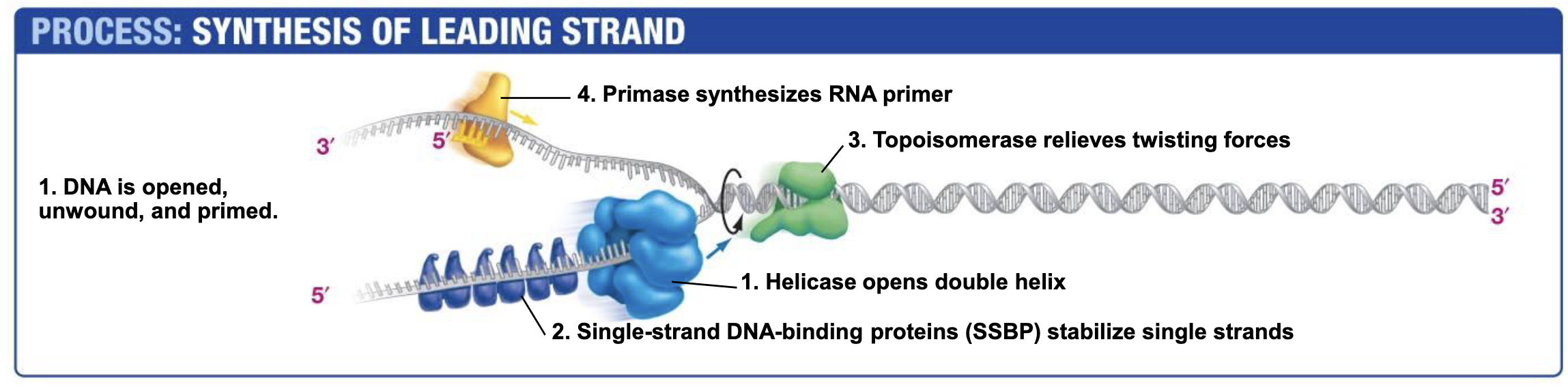
Primase (RNA Polymerases)
Catalyzes the synthesis of RNA primers that are required begin DNA synthesis.
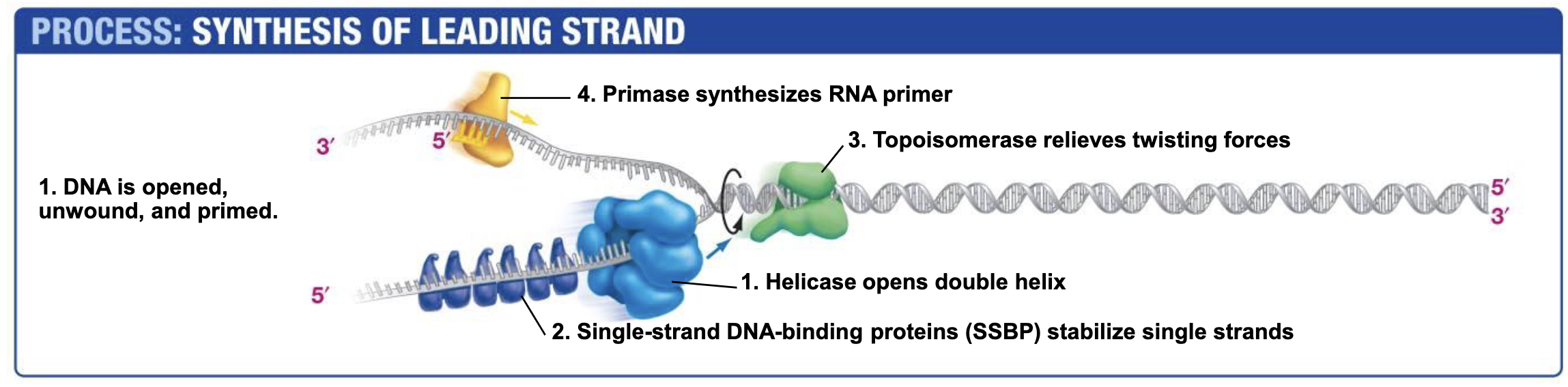
Why RNA Primers?
DNA cannot be synthesized unless there is a free 3’ OH present (But RNA can be synthesized regardless)
Why is DNA synthesized 5’ → 3’
DNA Polymerase III can only synthesize 5’ → 3’ (IDK WHY)
DNA Polymerase III/Sliding Clamp
Synthesizes DNA 5’ → 3’
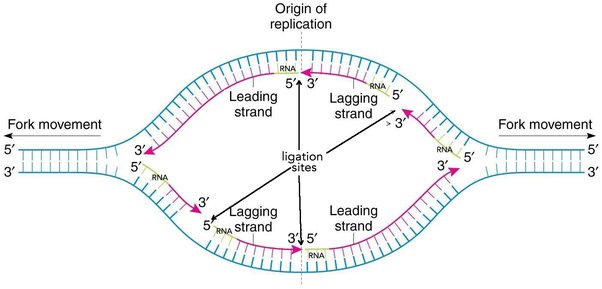
Lagging Strand
DNA is synthesized in the direction of helicase (direction that fork opens), which causes a discontinuous DNA synthesis on a strand.
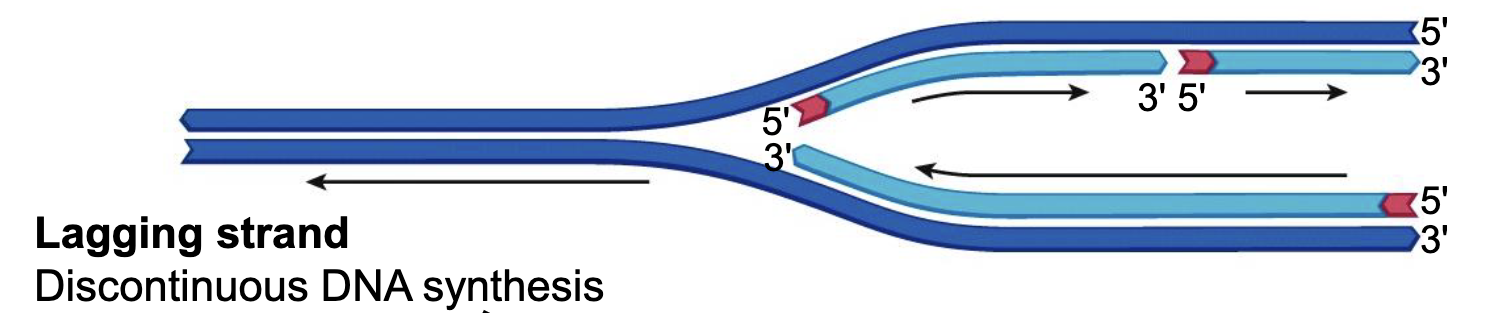
Leading Strand
The strand where there is continuous DNA synthesis. (Bottom in image)
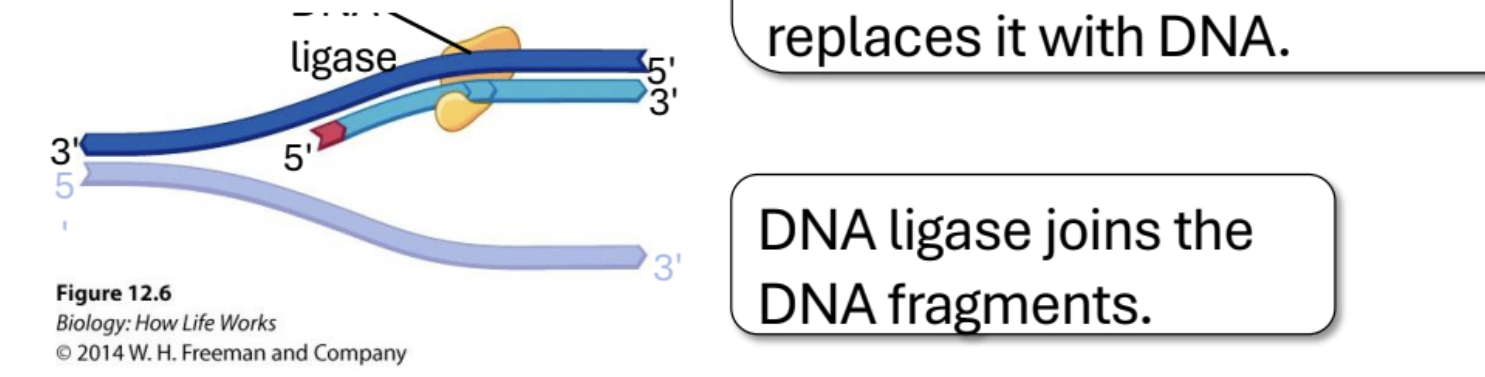
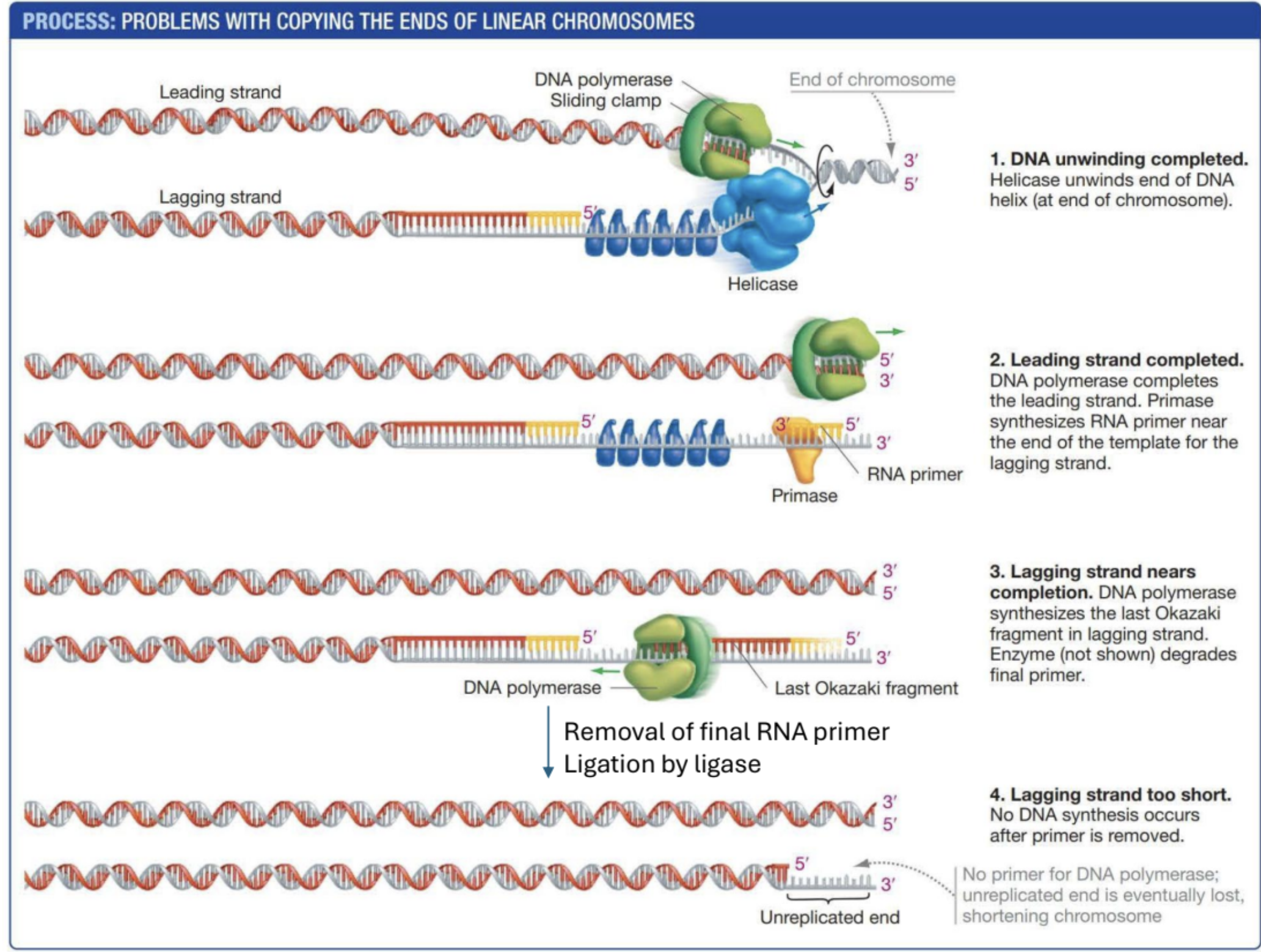
DNA Polymerase I
Removes RNA primer and replaces it with DNA (Important for lagging strand). After the second okazaki fragment is added, polymerase 1 replaces the RNA primer with DNA.

Ligase
Joins DNA fragments after Polymerase I
Draw a replication bubble. Label 5’ and 3’ ends, lagging leading stand, and direction that helicase opens the strand.
Describe the end copying problem. Draw an illustration if possible
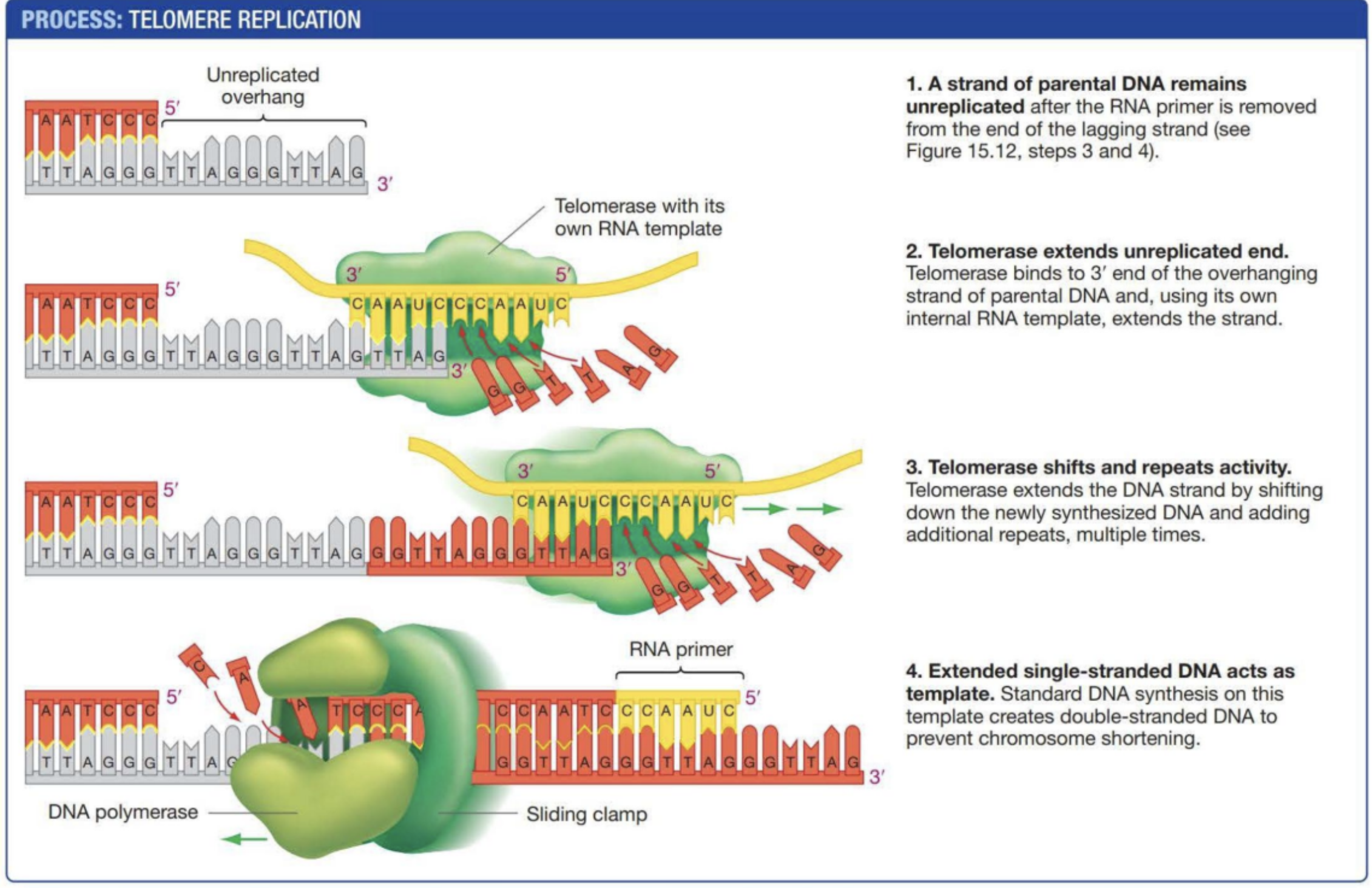
Telomeres
Repetitive strands of DNA at the end of chromosomes to prevent damage of chromosomes (Damage that comes from end copying probelm)
Telomerase
An enzyme with an internal RNA template to synthesize telomeres on a strand of DNA. DNA polymerase then synthesizes the complimentary strand to form a base paired double helix. Not all cells have this so DNA will eventually shorten causing genetic damage.
Why don’t bacteria
have telomerase/telomeres in general?
Bacteria have circular chromosomes → No end shortening problem
Mutations in somatic cells result in clones of [blank] cells surrounded by non-[blank] cells
Mutant, Nonmutant
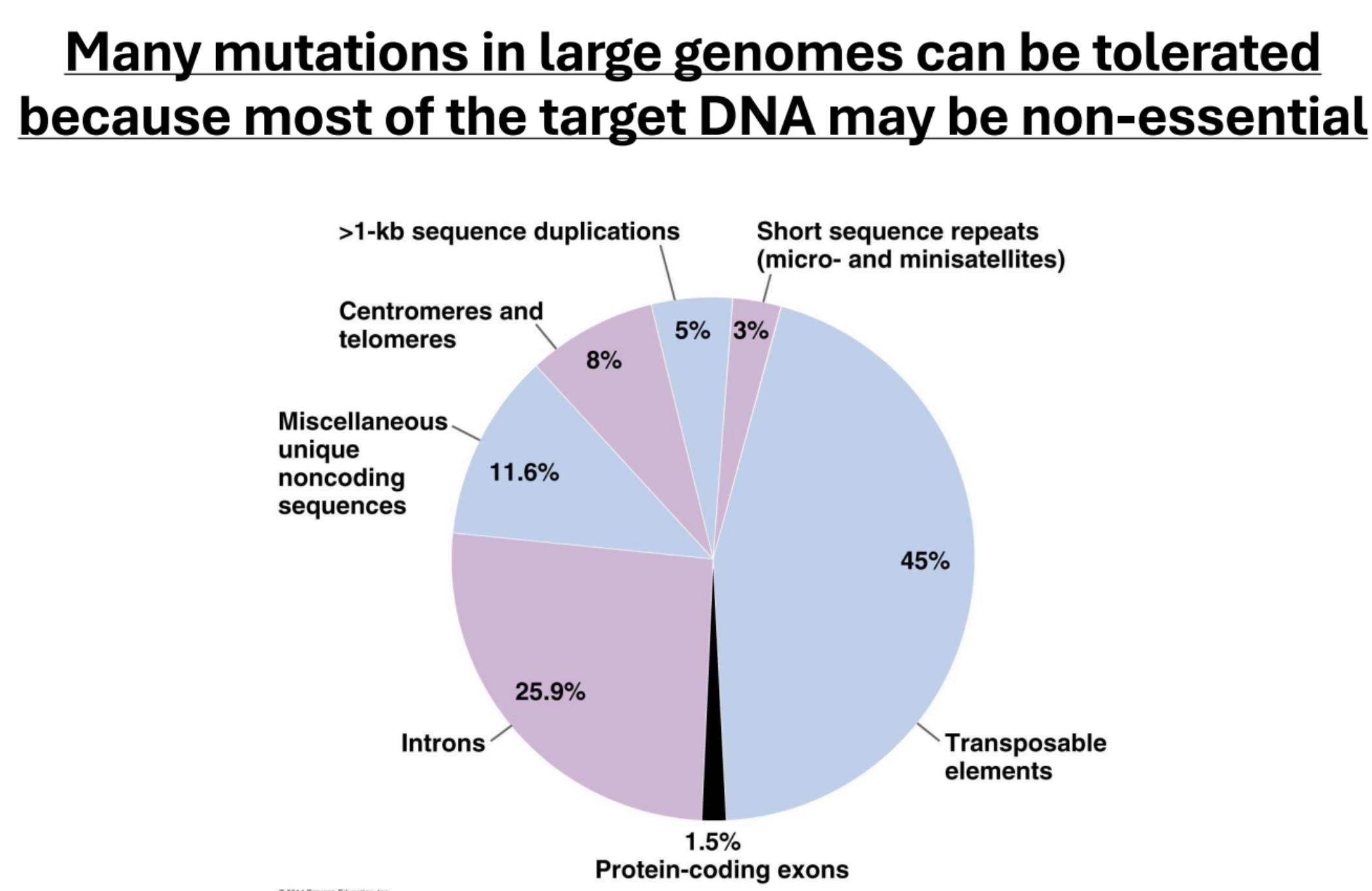
Why can many mutations in large genomes can be tolerated?
Because most of the target DNA may be non-essential
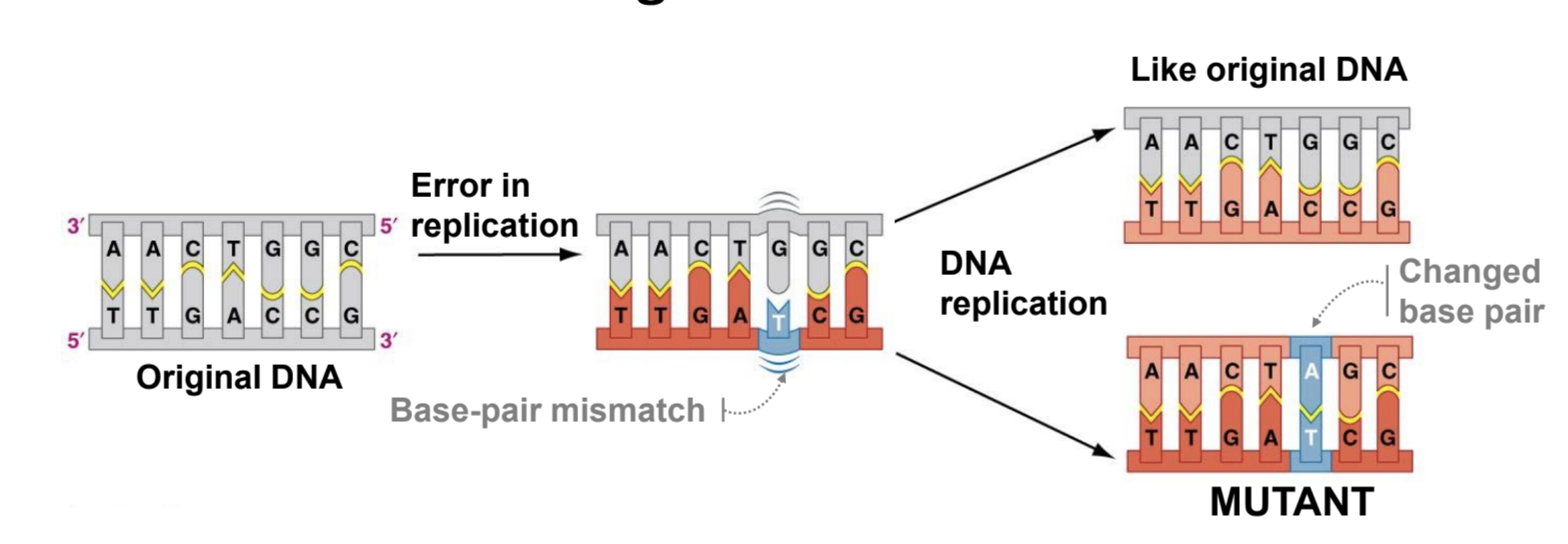
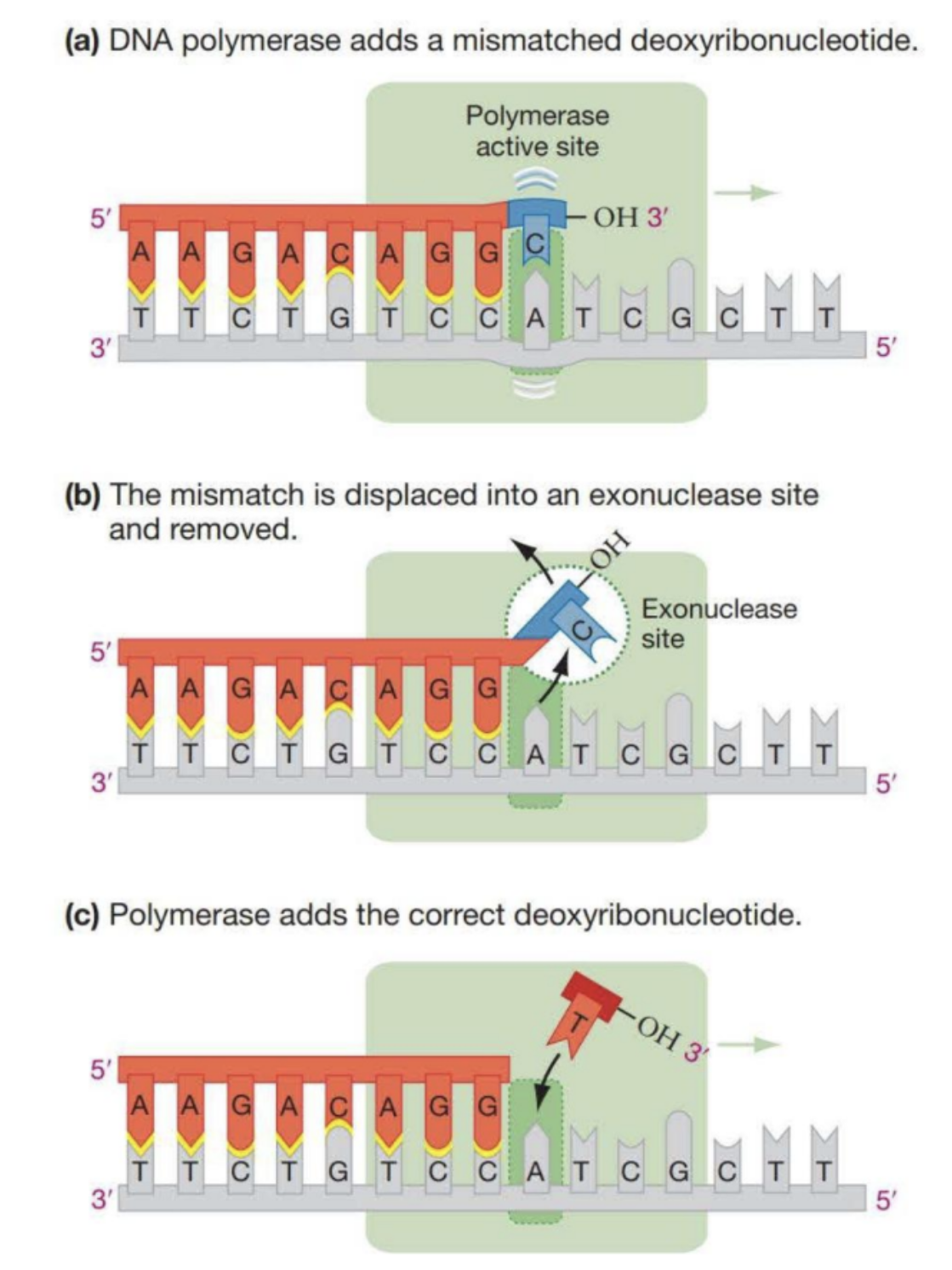
Polymerase “Proof Reading”
1/100,000 times DNA polymerase adds a wrong base. It can detect that there is a wrong base and moves the base to the exonuclease site. From there the correct base is added.
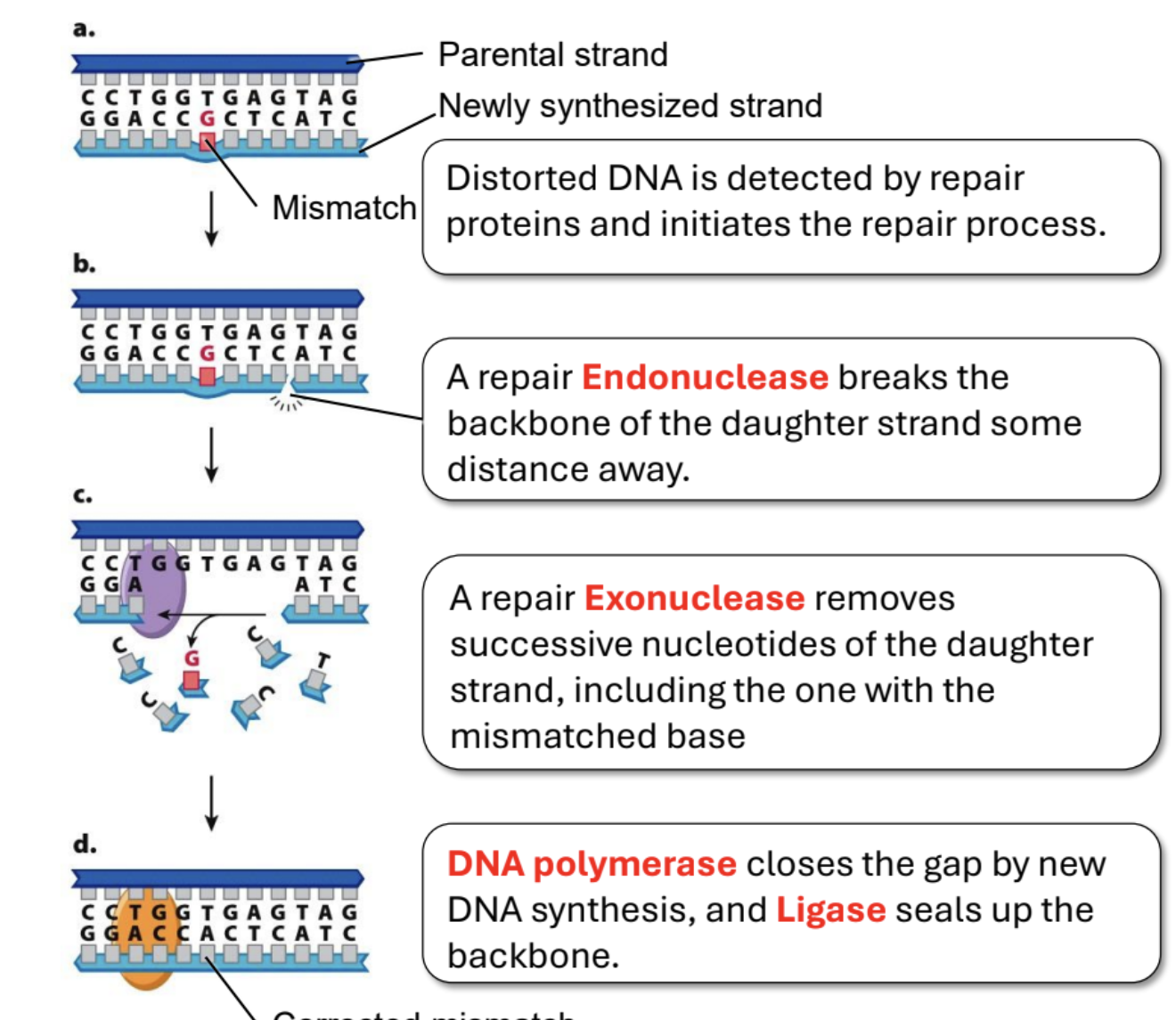
Give an overview of Mismatch Repair
Mismatched base pairs are detected by repair proteins which initiates the process. Endonuclease cuts the DNA some distance away from the mistake on the daughter strand. Then exonuclease begins removing all nucleotides that follow. Then polymerase synthesizes the correct DNA (there’s a free 3’ OH), while ligase connects the synthesized strand and the non-mismatched part of the daughter strand.
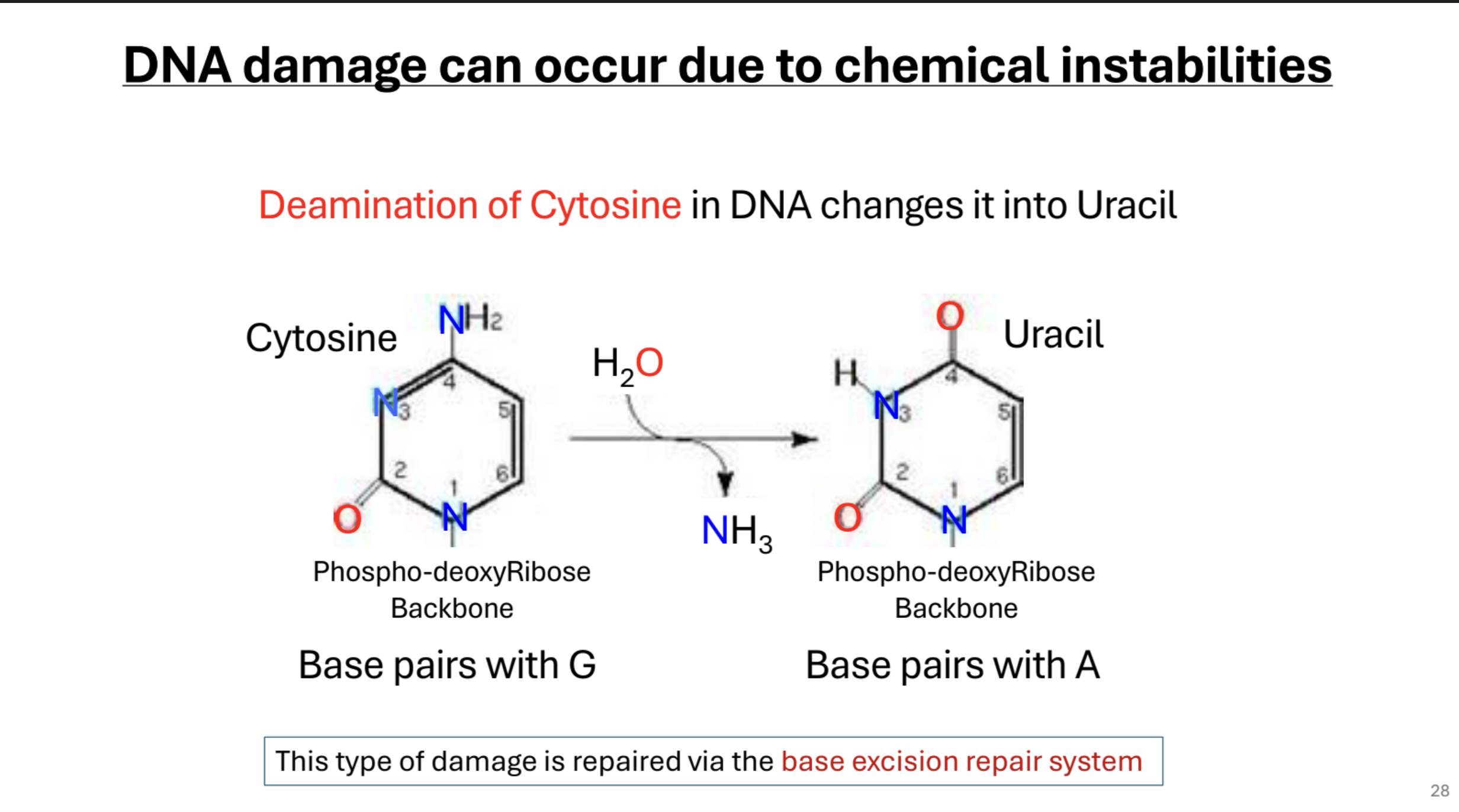
Deamination of Cytosine. What mechanism repairs this mutation?
DNA damage can be caused by chemical instabilities such as the deamination of Cytosine, which changes it to Uracil. This is fixed by base-excision repair system.
Describe UV DNA Damage. What mechanism fixes this kind of damage?
UV light can cause adjacent thymine base pairs to covalently bond (Thymine dimer), creating a kink in the sugar-phospate backbone of DNA. Nucleotide excision repair.
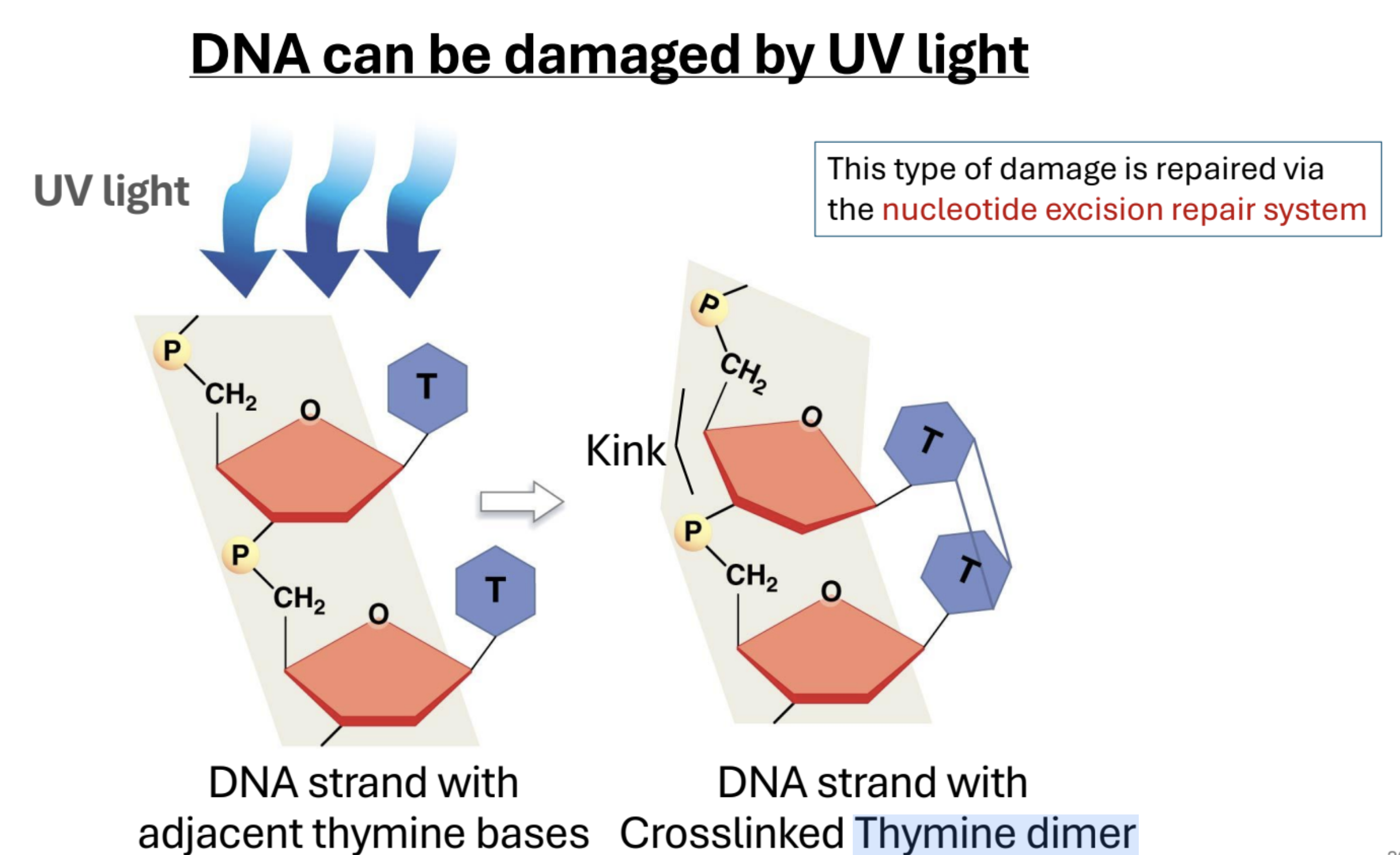
Describe nucleotide excision repair
Protein complex detects “kinked” thymines. DNA is nicked by enzymes on each side of the kink. Helicase unwinds and removes the region with damaged bases. DNA Polymerase reforms the DNA in the 5’ → 3’ direction using the uncut strand as a template. Ligase links the synthesized DNA back to its og strand.
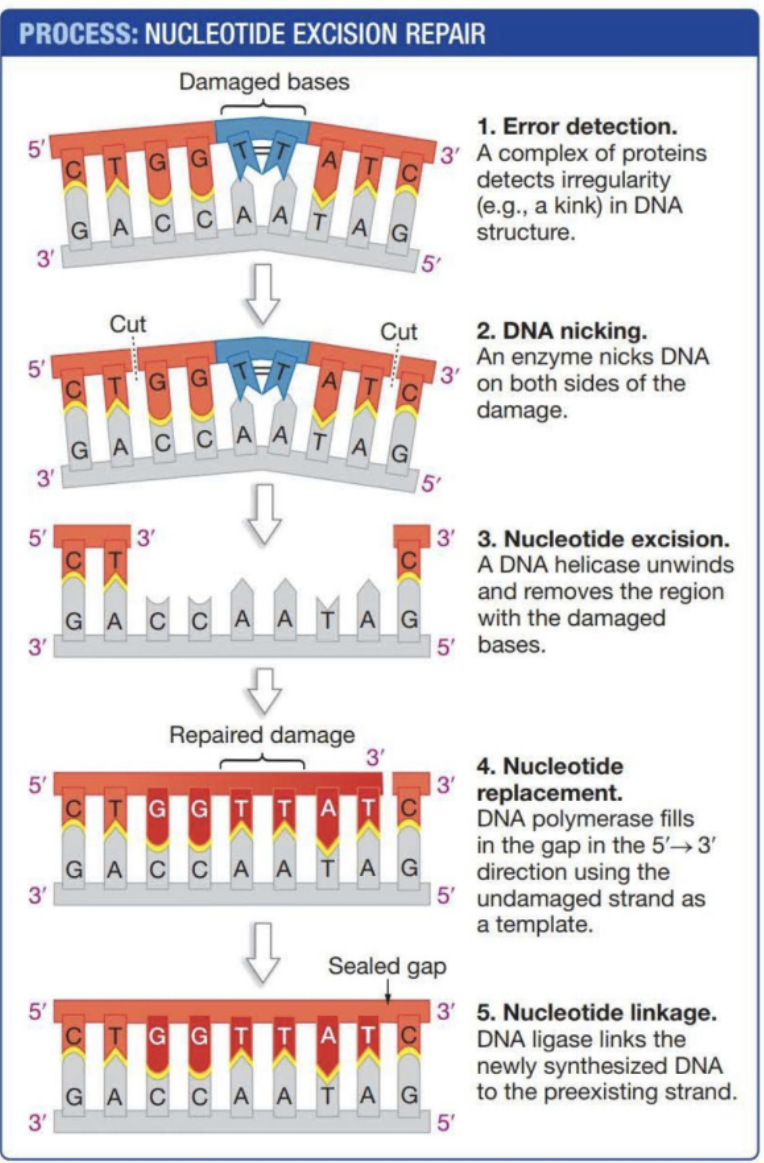
What are the four principle steps of most DNA repair mechanisms
Detect
Remove
Replace
Rejoin
Hershey-Chase Experiment
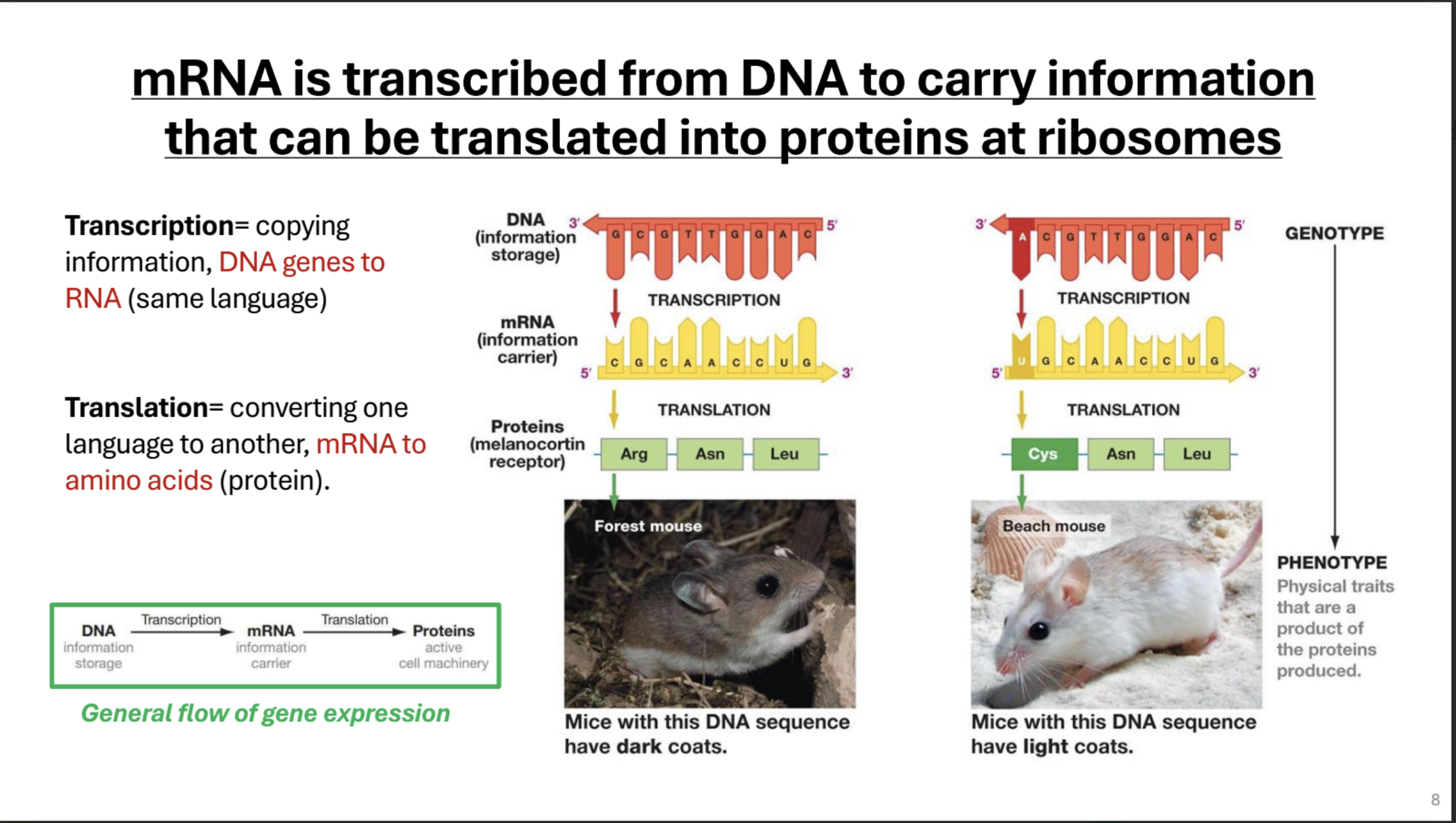
Describe the central dogma briefly
(Genotype) DNA stores genetic info
translation → mRNA which contains carries genetic info → Proteins (Cell machinery that creates phenotype)
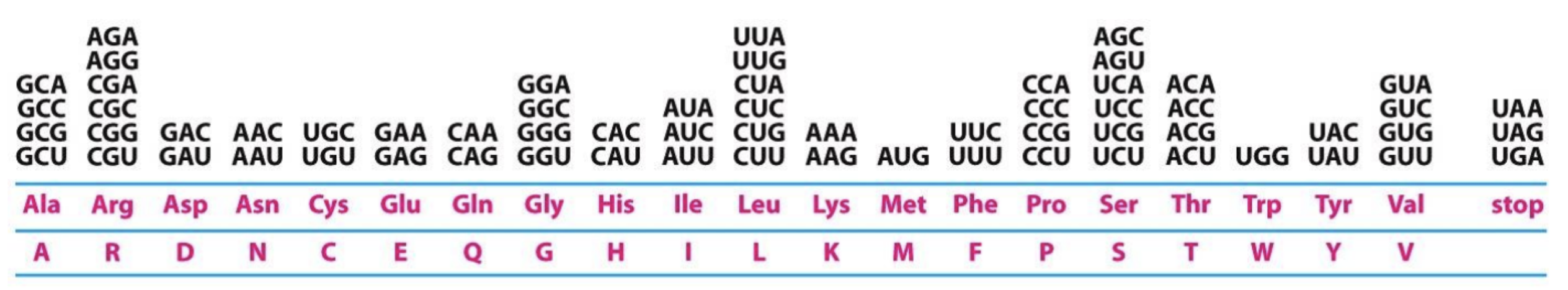
What are some important things about the genetic code?
It is redundant (degenerate): Multiple codons code for the same
No codon codes for more than one amino acid
It is nearly universal: with a few exceptions, all codons specify the same amino acid in all organisms
It is conservative: the first two bases of codons that specify the same amino acid will usually be the same.
Importance of reading frame.
The start codon AUG defines the reading frame for TRANSLATION. Since codons are read in threes, a change to the reading frame can be catastrophic, like a deletion or addition of a base pair.
Transcription and Translation do or do not start at the exact same place in a gene?
Do not
Name all types of mutations
Silent: Change in nucleotide sequence that does not alter amino acid. Usually change to third base pair
Missense: Change in nucleotide sequence that results in a change in the amino acid specified by codon.
Nonsence: Change in nucleotide sequence that results in a early stop codon
Frameshift: Addition or deletion of a nucleotide that changes
Why might a missense mutation not effect a proteins function? (THIS IS NOT SAYING MISSENSE HAS NO AFFECT ON PROTEIN FUNCTION, IT IS VERY LIKELY THAT IT CAN, TAKE SICKLE CELL FOR EXAMPLE)
If a polar/charged amino acid is replaced with a polar/charged amino acid, there might not be a significant change in tertiary structure. Same goes for hydrophobic proteins.
Describe how DNA is transcribed into RNA briefly.
RNA is synthesized from DNA template in the 5 → 3 direction, in the direction the DNA opens.
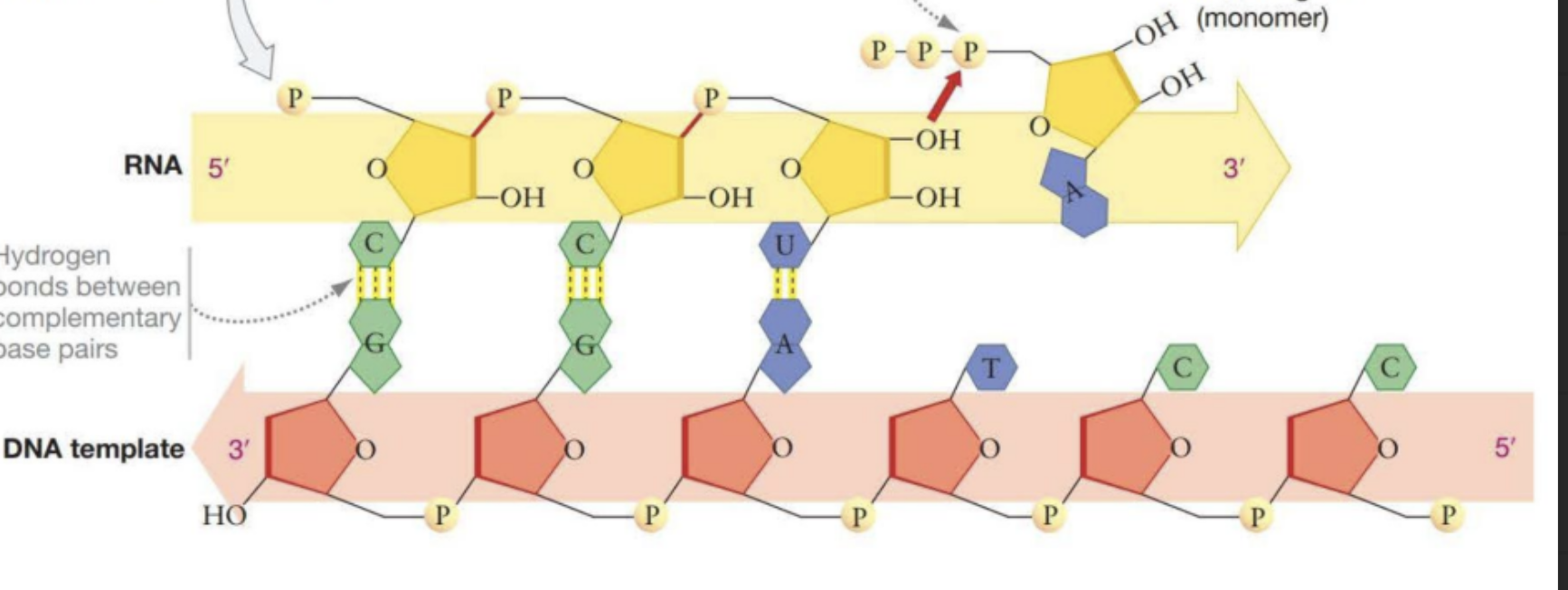
Template Strand
The strand of DNA on which RNA is synthesized. Complimentary to RNA
Coding Strand
The strand of DNAon which RNA is not synthesized. Matches RNA strand but with Ts instead of Us.
Draw a quick picture of RNA polymerase moving along a gene.
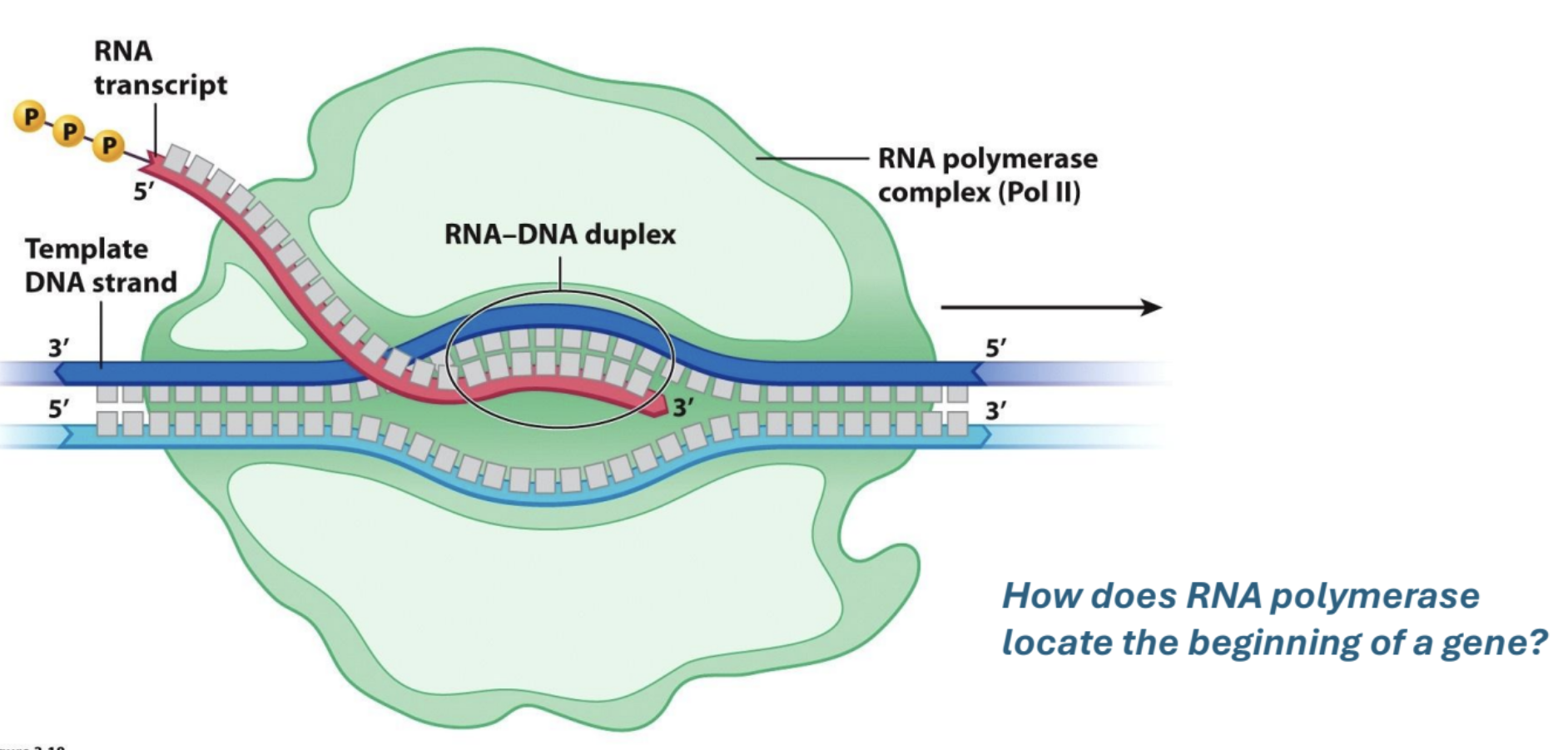
How does RNA polymerase locate the start of a gene?
Promoters
Transcription starts at ___ sequence and ends at ___ terminator sequence
promoter, terminator.
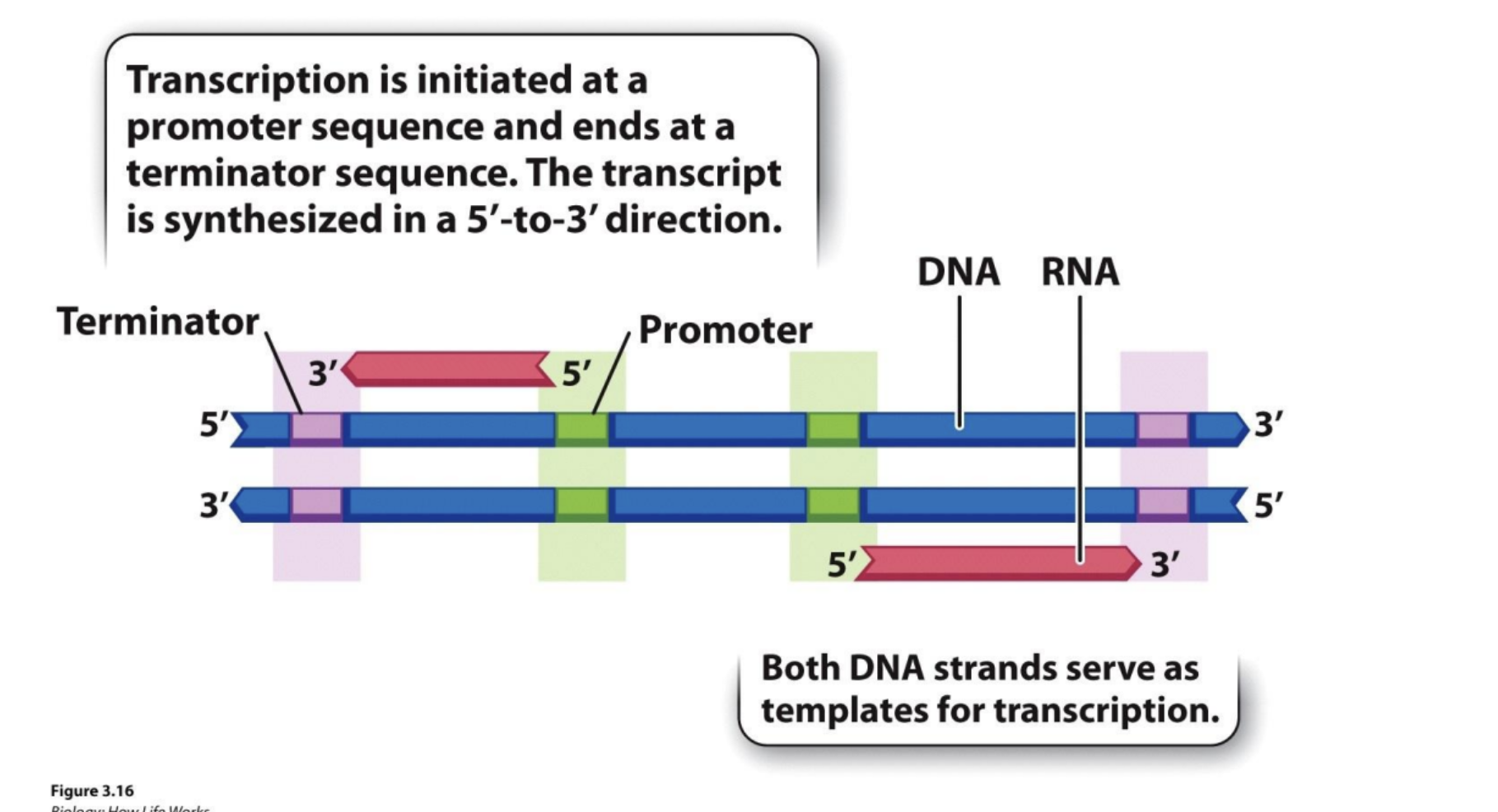
In prokaryotes, how does RNA polymerase orient itself?
Sigma
What does sigma recognize and bind to?
-35 box, -10 box in DNA promoter sequence (-35 meaning 35 bases away from start of transcription, -10 etc.) RNA polymerase binds to sigma before sigma binds to these boxes
How does sigma factor recognize DNA sequences?
Forms base specific hydrogen bonds with edges of base pairs.
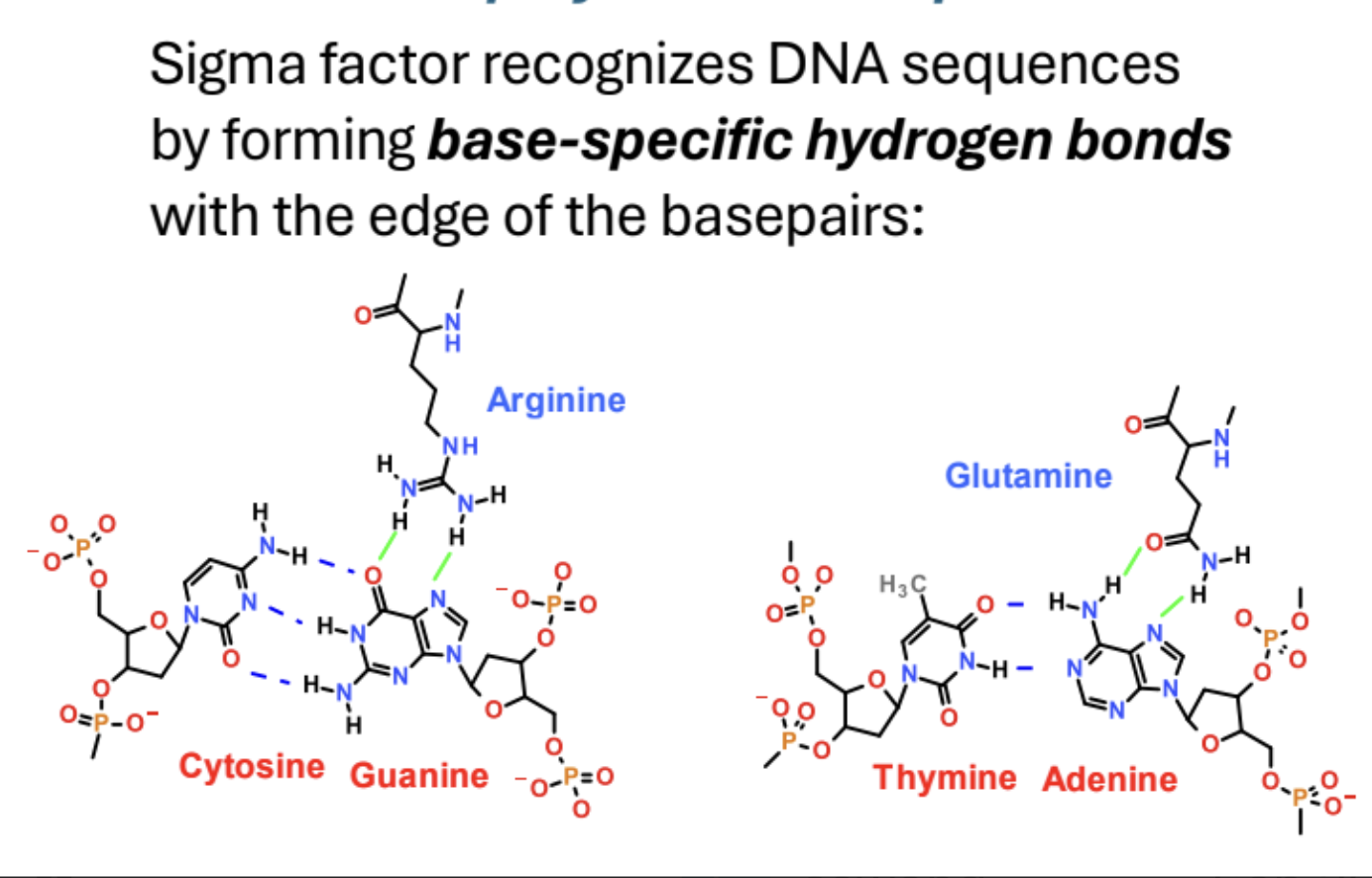
Why does sigma binding orient RNA polymerase?
Sigma binding orients RNA Pol. on DNA because it can only bind to both the -35 box and -10 box in one orientation
Prokaryotic transcription – Overview of Steps
Initiation: promoter binding
Strand separation, Primer synthesis
Elongation
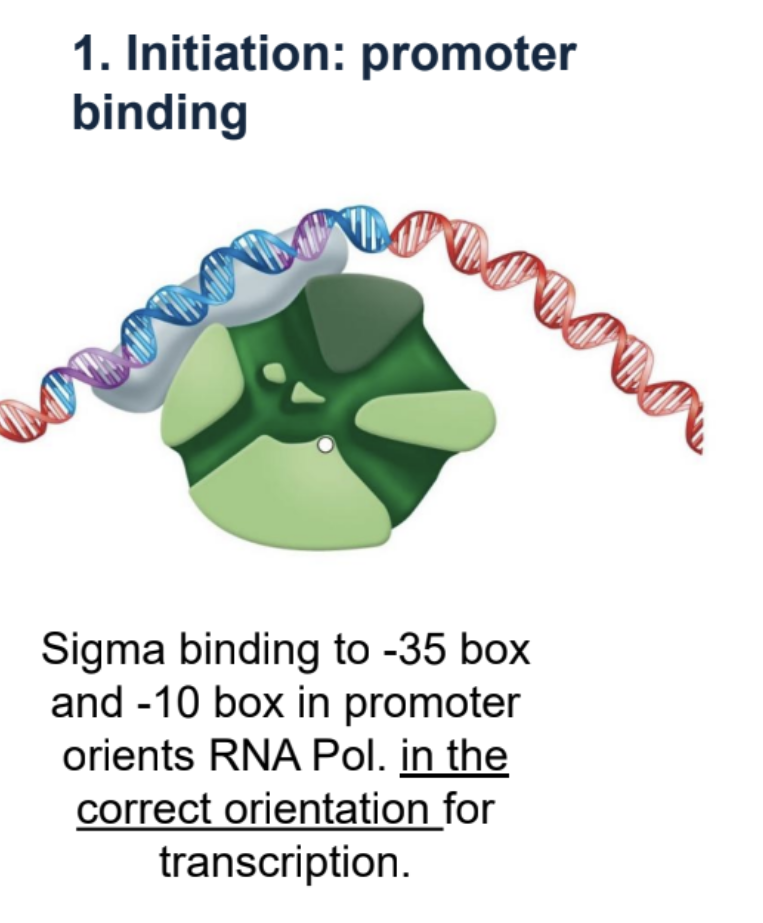
Prokaryotic transcription — Initiation: promoter binding
Sigma binds to -35 box and -10 box which orients RNA Pol.
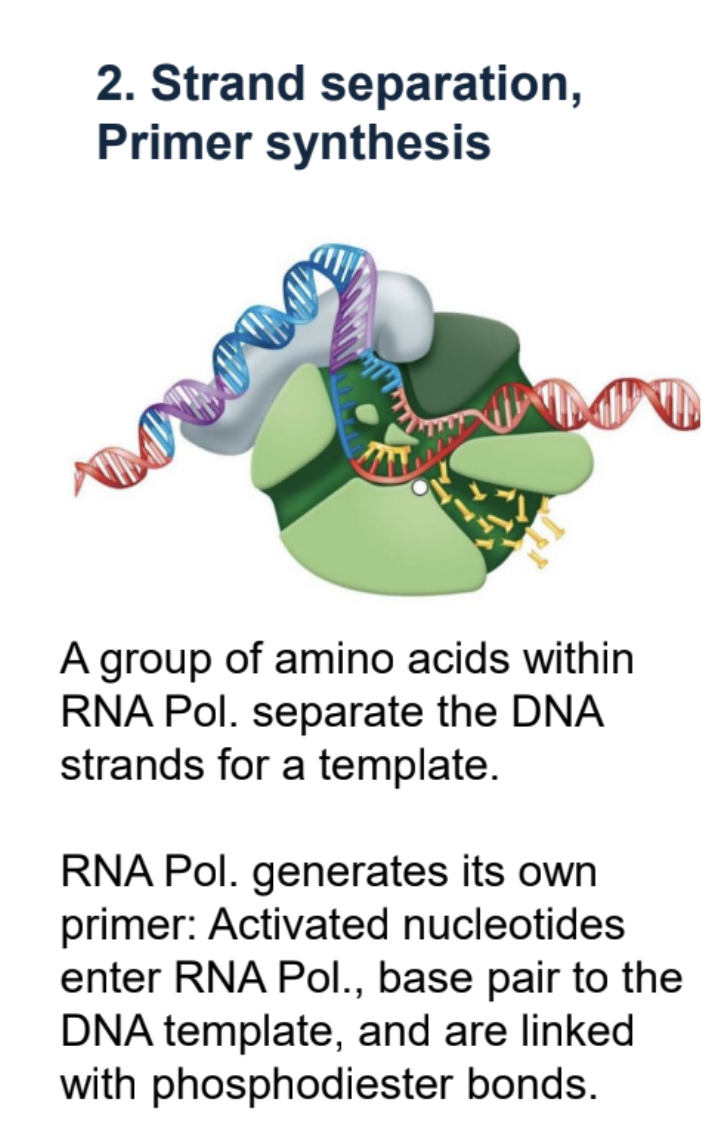
Prokaryotic Transcription — Strand Separation and primer synthesis.
Amino acids within RNA Pol. split DNA into template and coding strand. RNA pol. creates its own primer. Activated nucleotides enter RNA pol. and base pair with DNA template, joined by phospohodiester bonds.
Prokaryotic Transcription — Elongation
Sigma leaves and RNA pol. keeps transcribing 5’ to 3’.
Termination of Prokaryotic Transcription — Overview.
Hairpin formation
Release of RNA
Harpin formation
The termination signal codes for RNA that forms a hairpin loop by intra-RNA base pairing.
Release of RNA
The RNA hairpin causes the RNA strand to separate from the RNA polymerase, terminating transcription.
Things to note about eukaryotic transcription
In Eukaryotes, multiple types of transcription factors bind specific sequences of DNA to initiate transcription: Basal transcription factors, promoters, promoter proximal elements, enhancers (sequence), activator
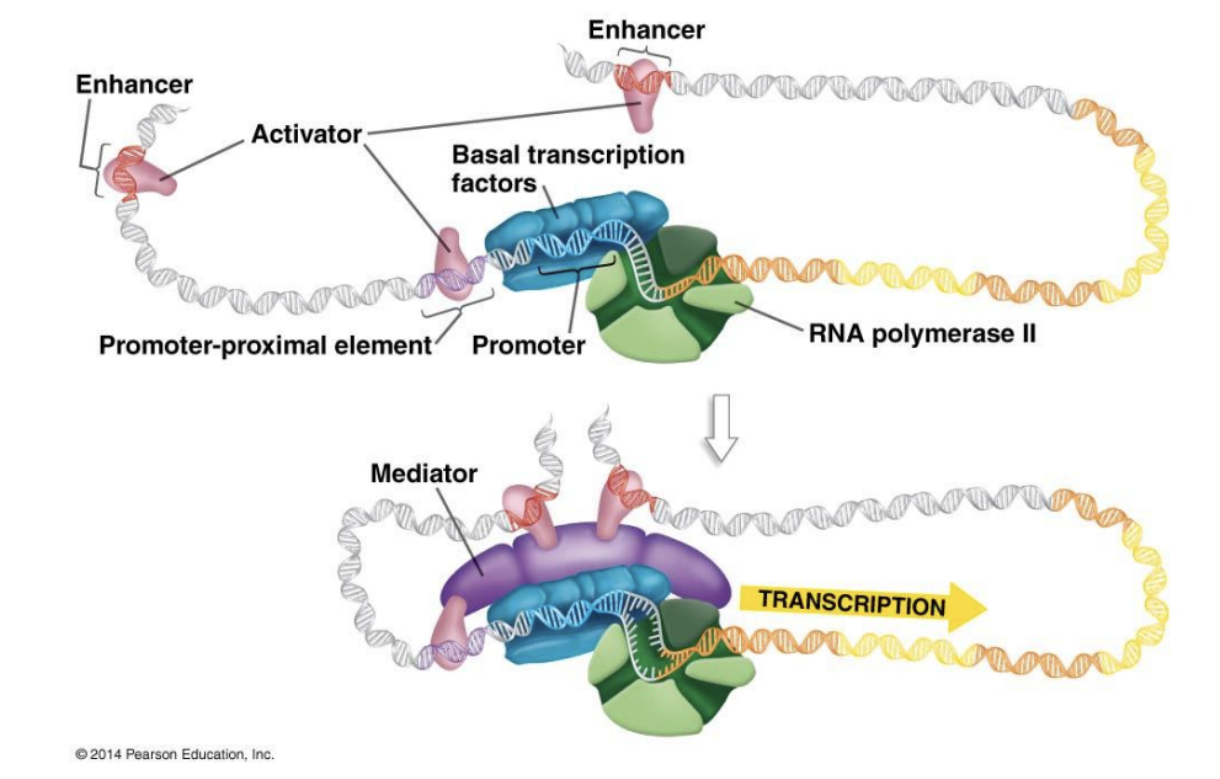
Basal Transcription Factors
Help initiate transcription at promoter. Orients RNA Pol. 2.
Alternative splicing
RNA in different cells can be spliced differently to create different proteins.
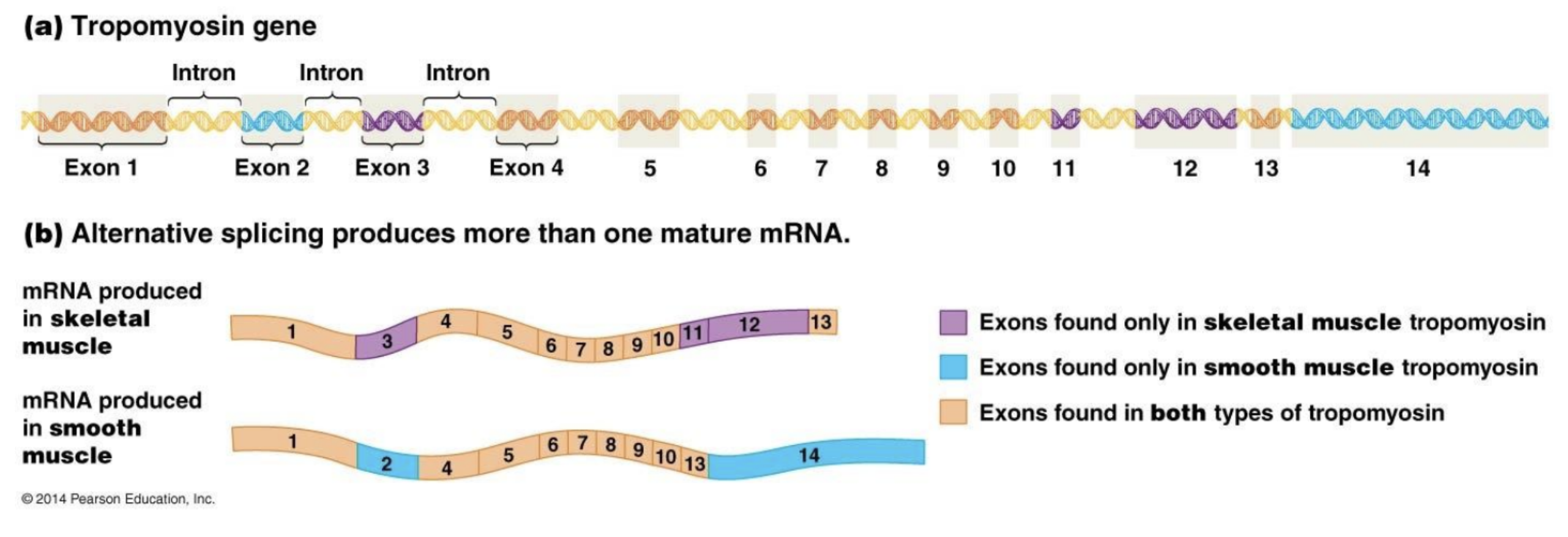
What must be removed from eukaryotic RNA before translation?
Introns
Enhancer
Sequences very far from start of gene in Eukaryotic DNA.
Activators
Bind to enhancer and make transcription more effective.
5’ Cap
element on 5’ end of mRNA in EUKARYOTES consisting of three phosphate groups and 7-methyl-guanosine.
Poly A tail
Sequence of 150-200 Adenine on 3’ end of mRNA in EUKARYOTES.
Purpose of 5’ cap and poly A-tail
Stabilize mRNA and allow it to be exported from the Nucleus
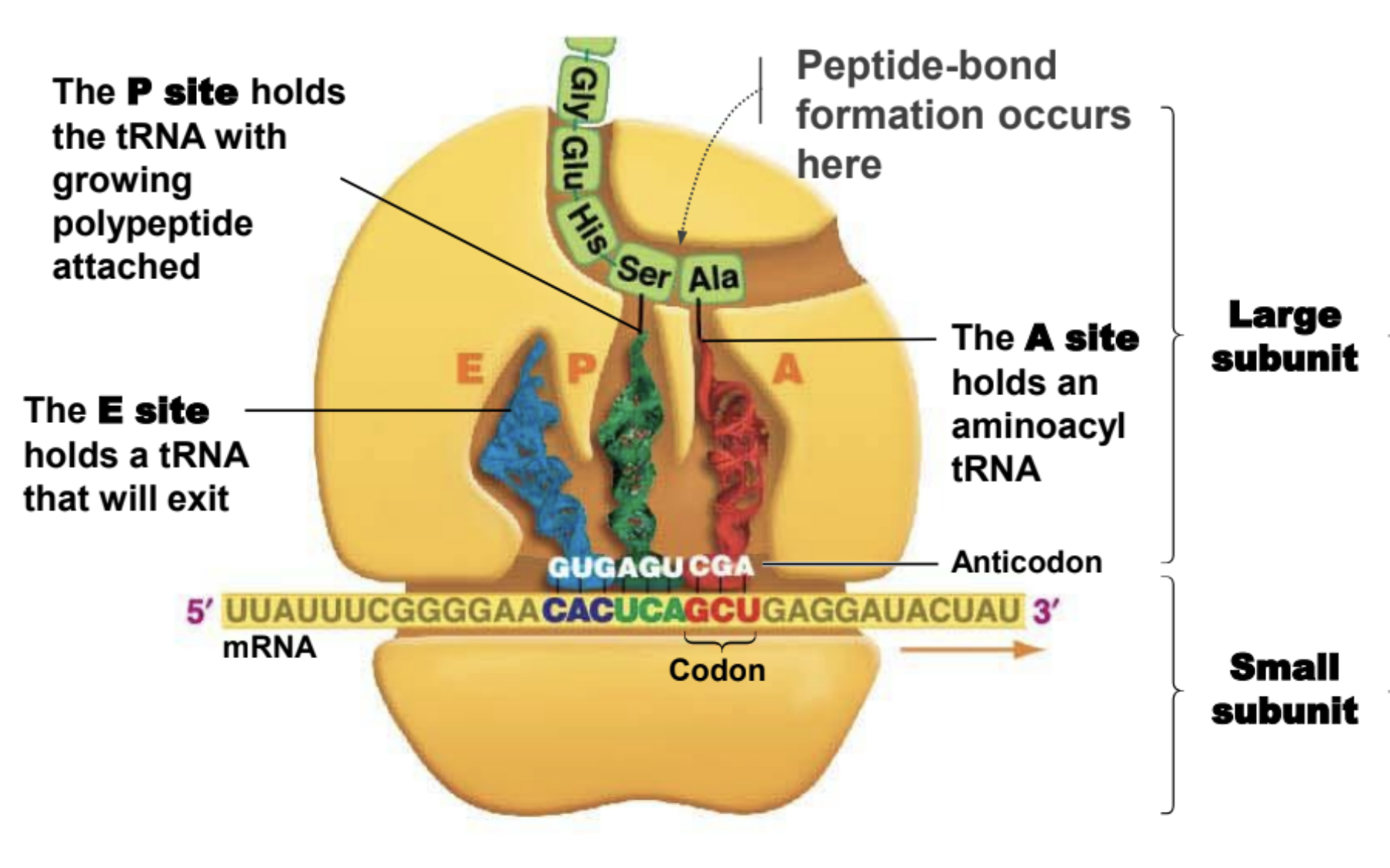
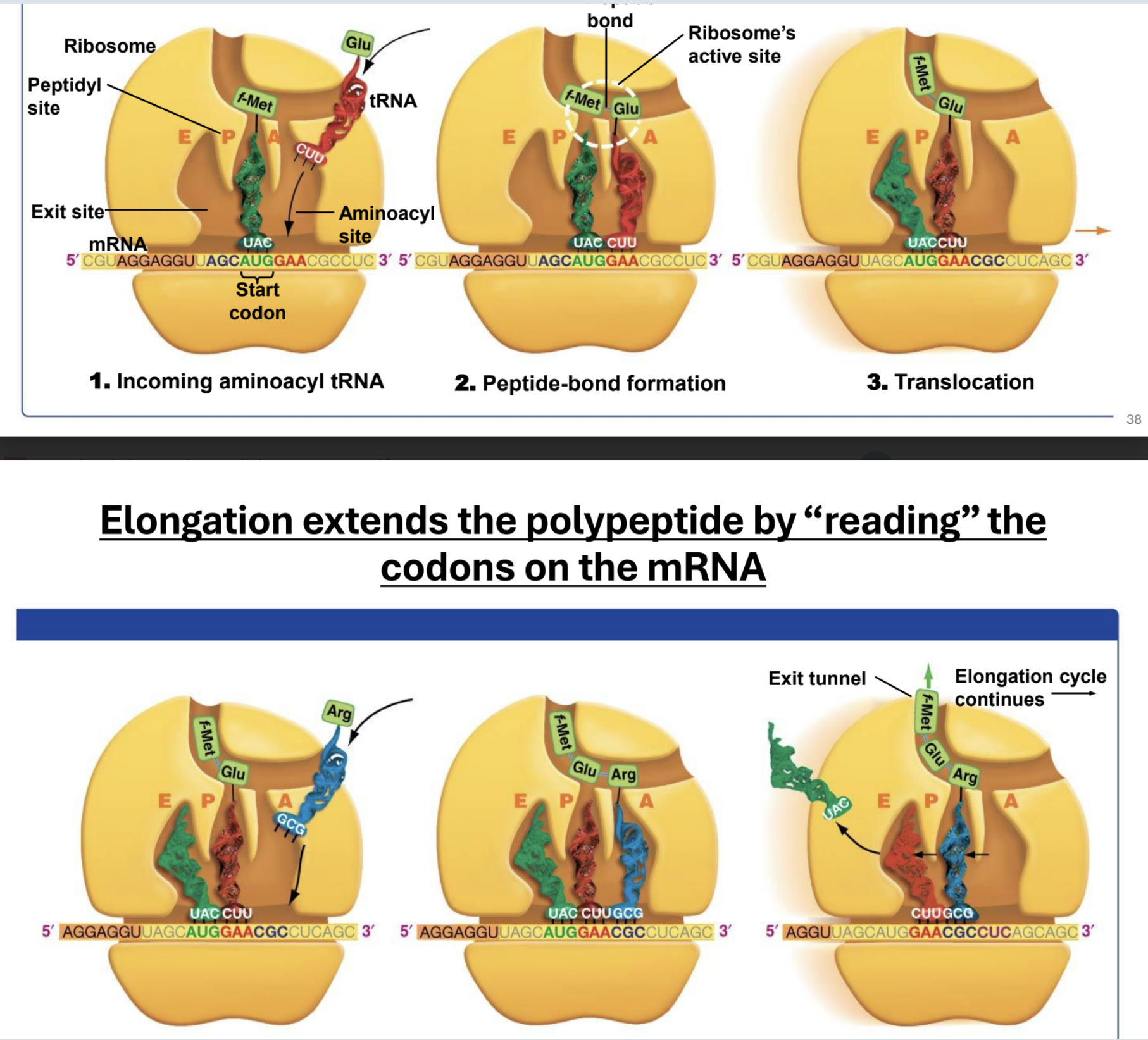
Important facts about ribosomes
They are made of rRNA.
Small subunit holds mRNA in place
The large subunit is where peptide-bond formation takes place
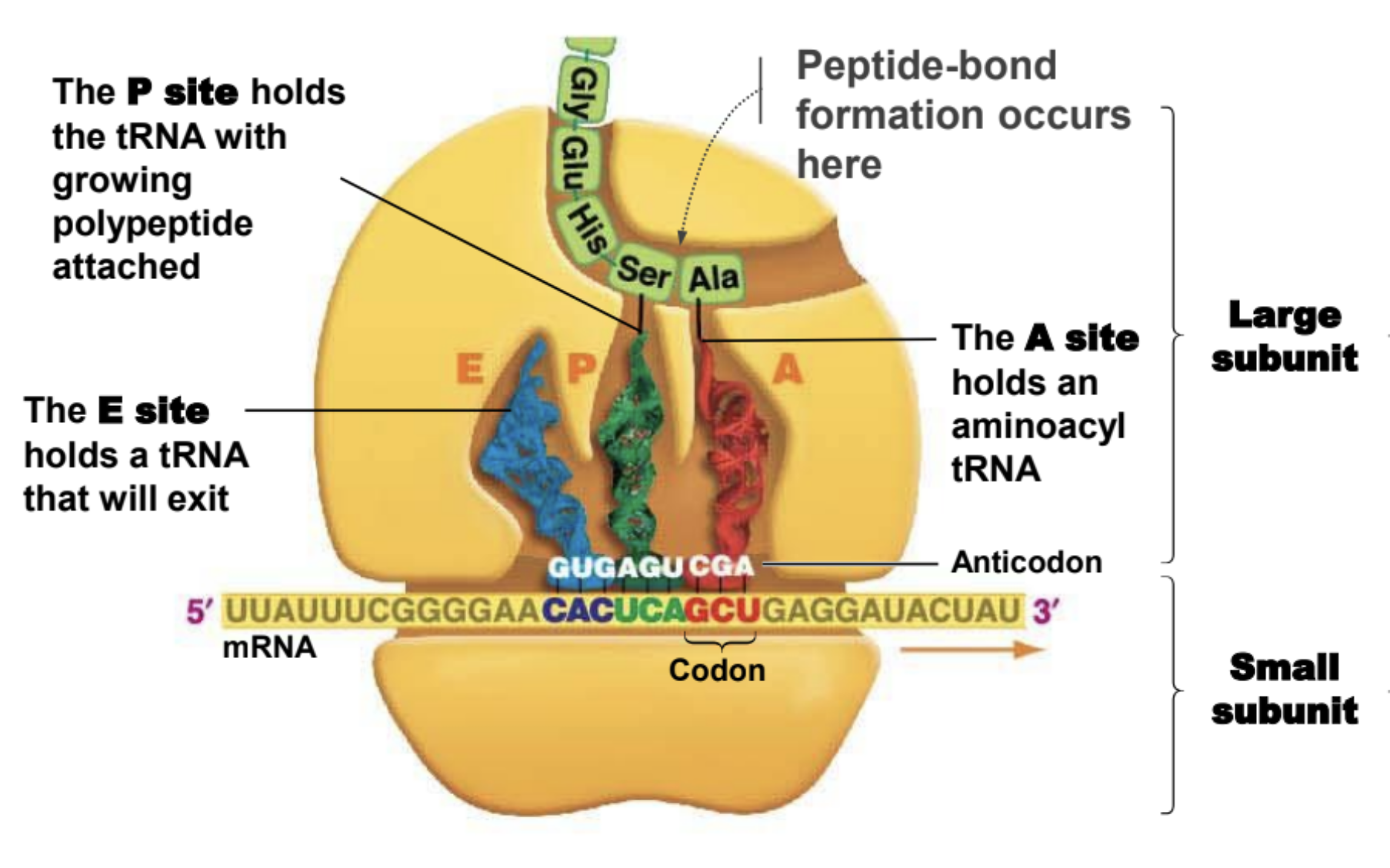
A site of ribosome
Hold entering amino acyl tRNA
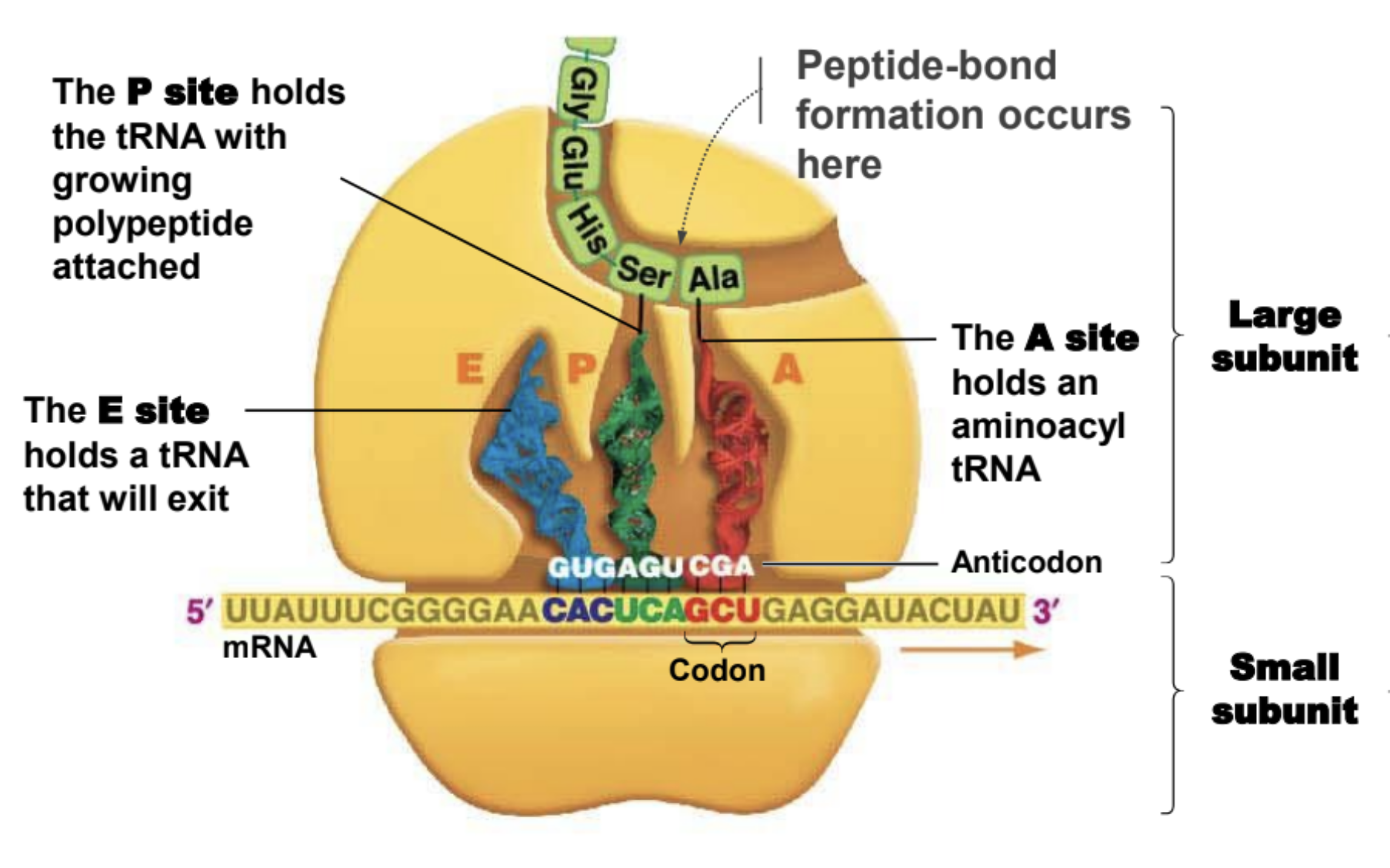
P site of ribosome
Holds tRNA with amino acid attached to polypeptide chain
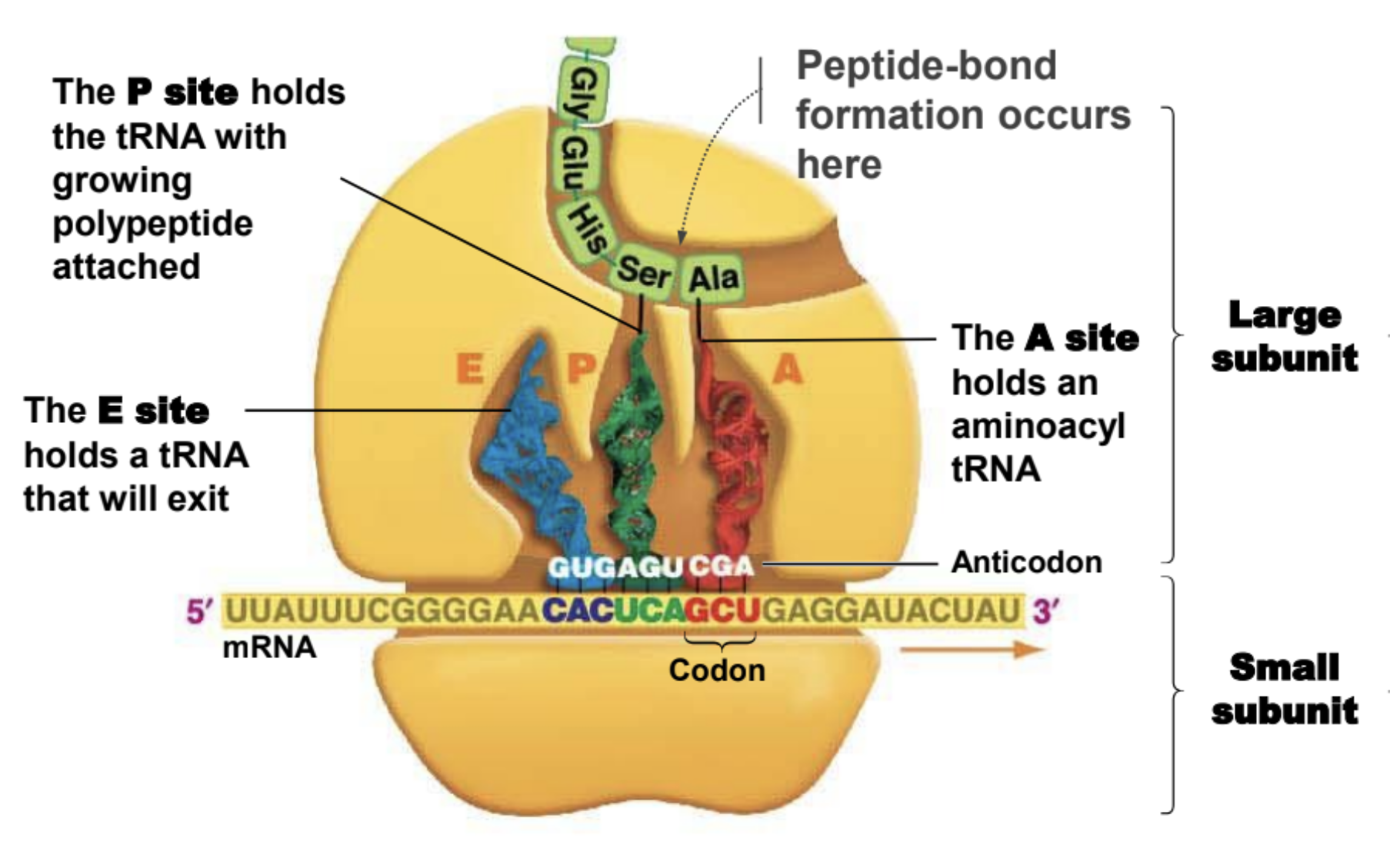
E site of a ribosome
Holds tRNA that will exit ribosome
Eukaryotes keep transcription and translation _——-
Separate. DNA and mRNA transcription in nucleus and translation in cytoplasm
Describe transcription and translation in Prokaryotes
Transcription and translation happen simultaneously. Multiple ribosomes can attach to a string of mRNA so multiple peptides can be formed per mRNA (polycistronic, and opposed to eukaryotic translation which is monocistronic)
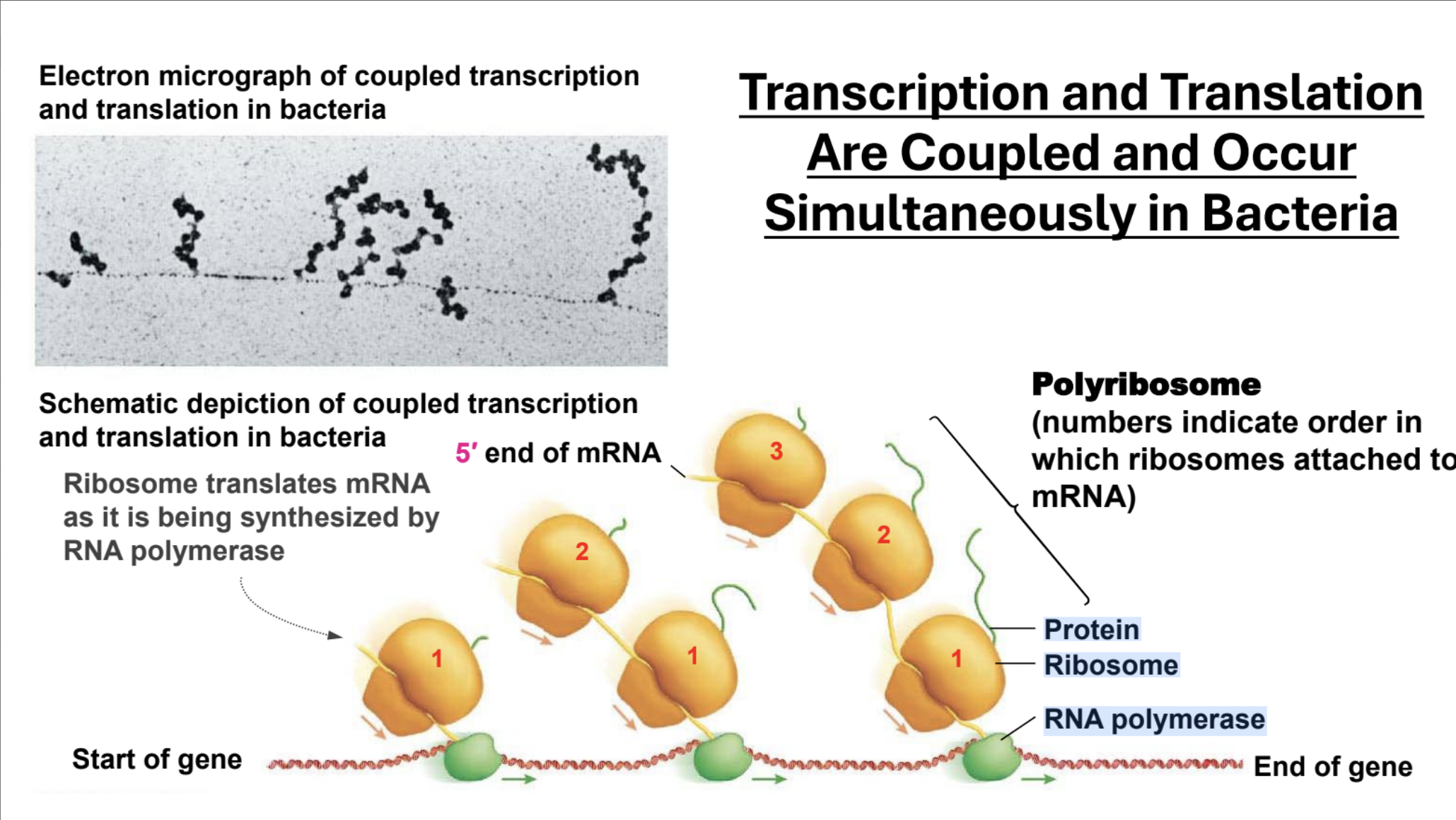
Polycistronic, Monocistronic
Polycistronic: The gene encodes multiple separate gene products. Monocistronic: The gene encodes one gene product.
Structure of tRNA
Made of RNA (obv)
CCA end with Amino Acid attached
Anticodon (Complimentary to 3 sequence codon, but not always the third pair)
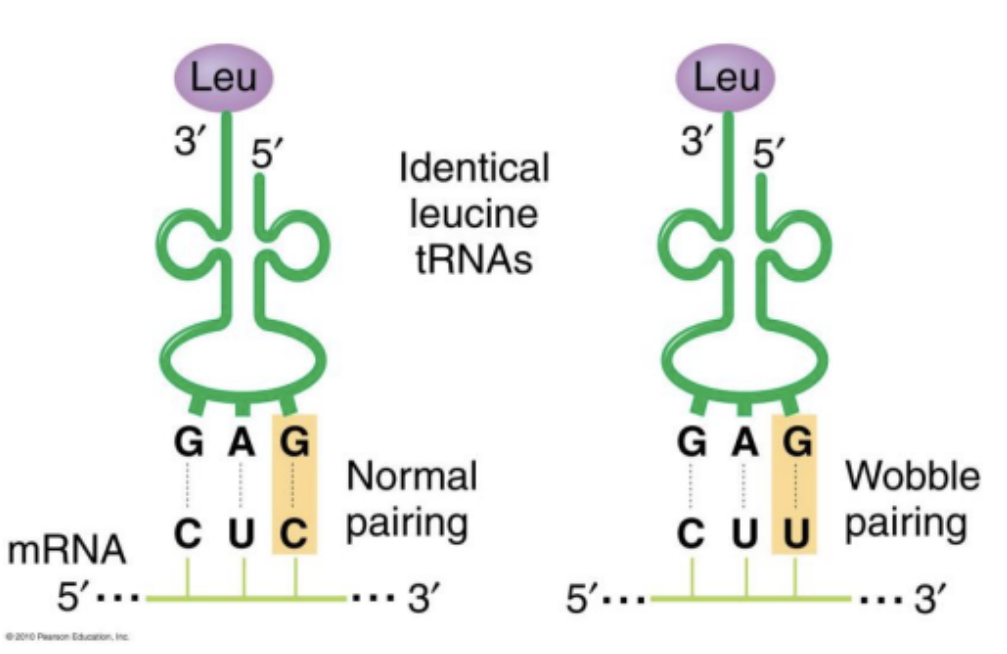
If an anti codon base pairs with a corresponding codon, how is codon specification degenerate?
Wobble pairings: Certain bases in the third position of tRNA anti-codons can bind to bases in a manner that does not match complementary pairing allowing one tRNA anti-codon to recognize more than one mRNA codon. This is called a wobble pairing.
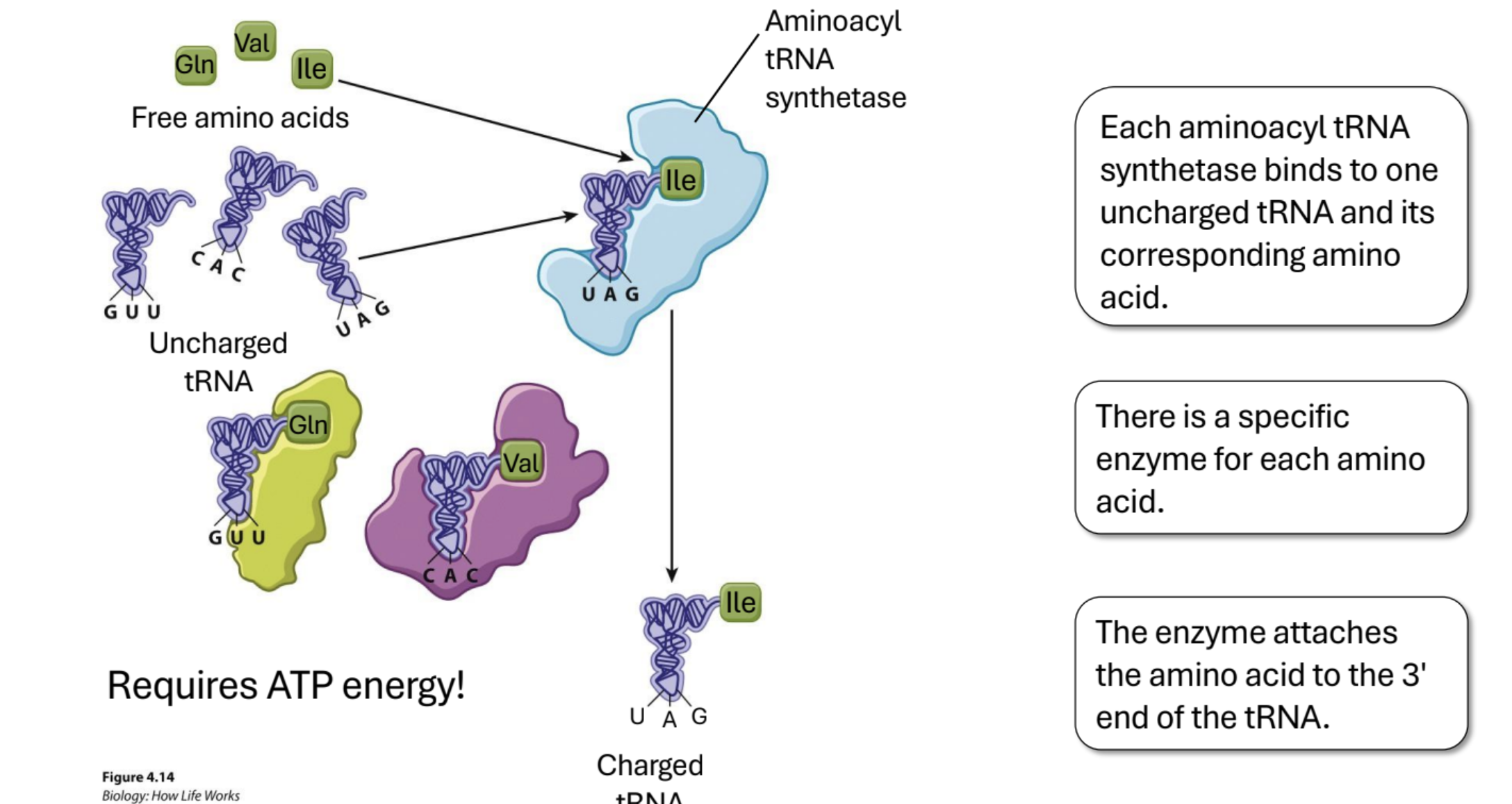
Charging of Amino Acids.
Enzyme (Aminoacyl tRNA synthetases) links AA with tRNA. There is a specific enzyme for each amino acid. Requires ATP. The enzyme attaches the amino acid to the 3' end of the tRNA.
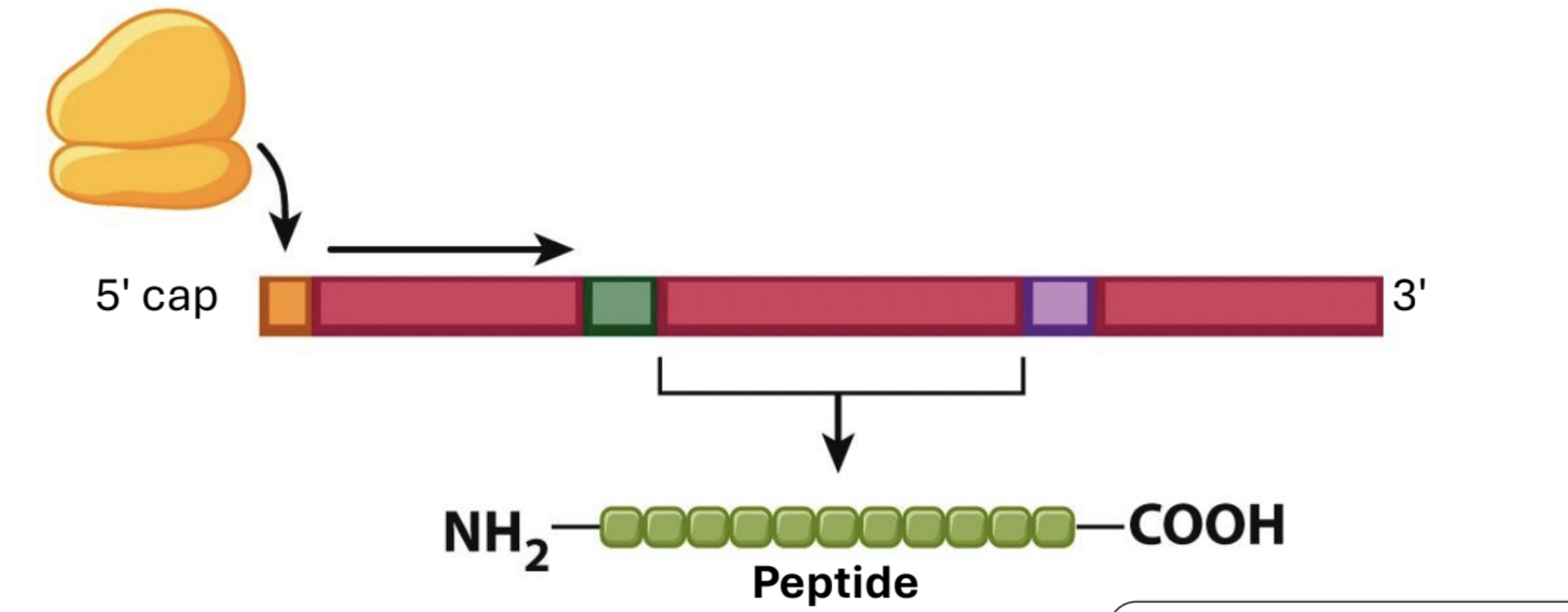
Translation Initiation in Eukaryotes…
is at the 5' cap, and the first AUG is the start codon
Translation in Prokaryotes is…
Translation initiation in Prokaryotes is at any Shine–Dalgarno sequence
Ribosome building peptide chain (just think about it or draw it)
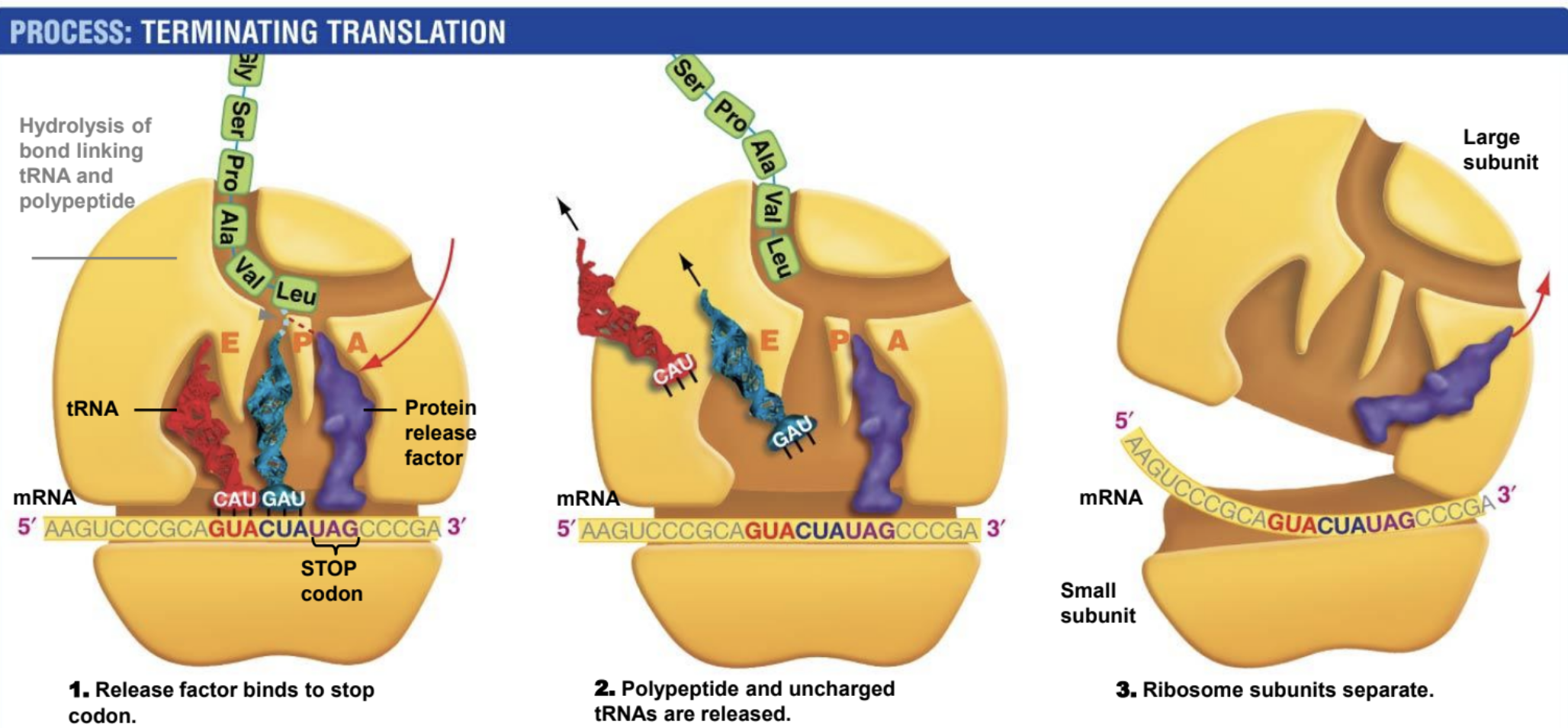
Termination (ending translation)
Protein release factor binds to stop codon. Polypeptide and uncharged tRNA are released. Ribosome subunit splits.
When does protein folding happen?
Along with translation.
What direction is a peptide built in?
N terminus to C terminus
