02 - Packaging & Organizing Genomes
1/65
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
66 Terms
In what ways are these genomes likely to differ or be similar?
Genomes are continually changing. What do we mean by change?
How/why does change happen? What could occur from parent to offspring/daughter cell?
Eukaryotes
Prokaryotic cells (Archaea and bacteria):
Human chromosome structure
are linear and we have 2 of each (23 pairs), meaning we are diploid
What can your cells do that a bacteria can do too?
does size correalate with number of genes
Genome size varies tremendously! But size does NOT necessarily correlate with number of genes (or chromosomes)
True or false: All eukaryotes have larger genomes than all prokaryotes
True or false: Compared to eukaryotes, the number of genes in prokaryotic genomes correlates more strongly with genome size
Rank the following in order of average increasing gene density: Prokaryotes, Mitochondria, Eukaryotes
What do you think this means about the composition of your genome?
• Non-coding DNA can make up a ____ of eukaryotic genomes
large %
Non-coding DNA is often ___
repetitive
From chromosomes to genes
Non -coding sequences can exist within genes
Once transcribed into RNA, introns are removed by the process of RNA splicing
The remaining protein - coding sequences are called exons
What do you notice about the spacing and sizes of genes and exons?
Packaging the eukaryotic genome
Genome must be packaged in order to fit in the nucleus:
Packaging must be compact but also accessible to enzymes and regulatory factors
DNA Supercoiling
Supercoiling is helpful to relieve the strain
• Increases stability
• Making DNA more compact
• Allows unwinding of sections
Why would we need to unwind (open) a section of DNA?
Positive supercoils →
helix is overwound
Negative supercoils →
helix is underwound
What might happen to a cell if too much supercoiling prevents helix opening?
Topoisomerases
are protein enzymes that regulate/relieve supercoiling
chromatin
Eukaryotic DNA associates with histones and other proteins to form chromatin
nucleosomes
Histones package DNA into repeating units called nucleosomes
• Repeating subunit of DNA + histones
• 8 histones proteins form a central core
• Octamer: two each of H2A, H2B, H3 and H4
• DNA is wound around the octamer core
Overall, packaging ratio of DNA in nucleosomes is
7:1
Histones
are highly conserved proteins rich in basic amino acids (Lys, Arg,) resulting in a high positive charge which interacts easily interact with the negative (-) charge of DNA
Where does the negative charge on DNA come from?
phosphate backbone
Histone dimer formation is mediated by
C-terminal domain histone fold
Each histone has an N-terminal histone tail that
protrudes outward and can be subject to covalent modifications
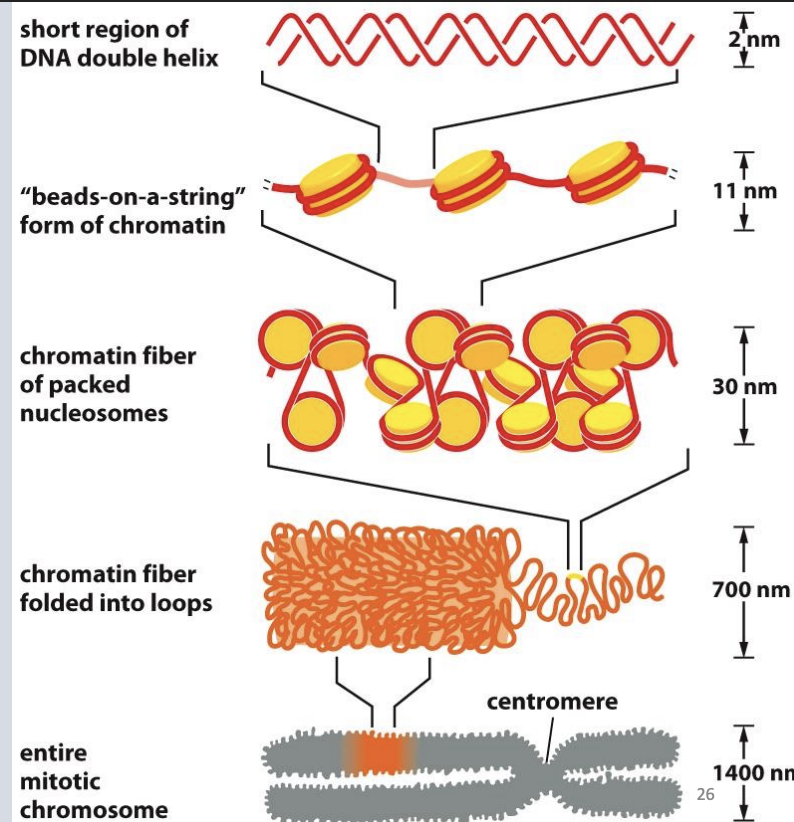
30 nm fiber
• Histone H1 (linker histone) binds linker DNA that connects one nucleosome to another
• Regulates how tightly these nucleosomes pack together
• Without histone H1, nucleosomes look like ‘beads on a string’
• Histone H1 can line up nucleosomes end-to-end into two stacks
• This “30 nm fiber” increases the DNA-packaging ratio 6-fold further (overall now ~40-fold)
– Looped domains
• 30 nm fiber may further be gathered into a series of large loops
• These loops are attached to protein scaffolds
Levels of chromatin organization
As cells prepare for mitosis, looped domains get further compacted
Euchromatin
DNA that is less compacted and functionally active (accessible for
protein binding and transcription). Genes here can be expressed
Heterochromatin
DNA that is highly compacted and has little to no functional activity
Types of Heterochromatin
Constitutive heterochromatin
Facultative heterochromatin
Constitutive heterochromatin
• Permanently silenced DNA
• Includes regions around telomeres and centromeres
• Contains DNA repeats and few genes
Facultative heterochromatin
• Inactivated during certain phases of organism’s life
Why is this necessary?
What do you think would happen if all your facultative
heterochromatin suddenly became euchromatin and vice versa?
Which of the following are true?
A) All DNA is facultative heterochromatin until it becomes constitutive heterochromatin
B) Centromeres are an example of constitutive heterochromatin
C) Genes in a region of euchromatin are more likely to be expressed than genes in a region of heterochromatin
D) After every round of cell division in females, one X chromosome is randomly selected for inactivation
E) X inactivation occurs in the female gamete (egg) before fertilization
Epigenetics
Covalent modifications (e.g. methylation) of DNA and histones
Influences heterochromatin vs euchromatin formation
Epigenetic regulation of gene expression can be inherited by daughter cells during cell division without changes to the sequence of nucleotides in DNA
Covalent modifications of histone tails can
disrupt or stabilize nucleosome assemblages
acetylation leads to what in histone code
more open structure and more transcription,
methylation causes what in histone code
compaction/repression
condensed nucleosomes
uncondensed nucleosomes
Histone acetyltransferases (HATs)
acetylate histone proteins by transferring acetyl group from acetyl-CoA to specific lysine residues.
Histone deacetylases (HDACs)
remove the acetyl group.
Histone methyltransferases (HMTs)
add methyl group(s) (1, 2, or 3) to lysine or arginine residues.
Histone demethylases
remove methyl groups
Most histone methylation contributes to
silencing (compacting) DNA
DNA methylation
DNA methylation works together with the histone code
Proteins that bind to methylated DNA can recruit enzymes involved in modifying histone tails
Methylation of cytosine can prevent proteins from binding to DNA sequences (often preventing initiating of transcription)
DNA methyltransferase
can add the methyl group to DNA at sites where a C is followed by a G (reading 5’ to 3’ = “CpG”) creating an epigenetic marker on the DNA
DNA demethylases
can remove these methyl groups.
DNA is methylated as it is
replicated so that methylation patterns can be passed on to daughter cells (The symmetry of CpG on both strands is critical for this!)
Maintaining epigenetic regulation
Silencing of genes by heterochromatin formation occurs in regions (“position effect”) and is maintained in replicated DNA
Does this mean that you inherit all your epigenetics from your parents?
Epigenetic regulation from gametes to offspring
Methylation patterns are copied to newly synthesized DNA and so can be inherited during cell division, but are mostly rewritten during initiation of offspring development
genomic imprinting
Some (very few) methylation patterns are passed on from parents to offspring (called genomic imprinting)
Maintaining heterochromatin after DNA replication
The epigenetic signals that regulate chromatin are propagated through space (along chromosome) and time (during cell division)!
Which of the enzymes mentioned so far are likely to contribute to creating a more condensed (heterochromatin) state? (select all that apply)
A) HATs (histone acetyltransferases)
B) Histone demethylases
C) HDACs (histone deacetylases)
D) Histone methyltransferases
E) DNA methyltransferases
Why doesn’t heterochromatin propagate across entire chromosomes?
Barrier DNA sequences can recruit protein complexes that block the spread of reader -writer complexes and separate chromatin into domains
Mechanisms of barrier action
Barrier proteins can bind to barrier sequences and create physical obstacles or actively recruit opposing chromatin modifying enzymes
Chromatin becomes separated into domains with different transcriptional activation/regulation.
If you wanted to understand how genomes evolve over time (many generations), would you look at the genetic code or the epigenetic modifications?
If you wanted to understand why your liver cell doesn’t behave the same way as a heart muscle cell, would you look at the genetic code or the epigenetic modifications?
Which of the following is TRUE?
A) You inherit all the epigenetic modifications in your genome from your parents
B) After each round of cell division, heterochromatin forms randomly in various regions of DNA
C) In your neurons and skin cells, the same set of genes are silenced by facultative heterochromatin formation
D) Your liver cell and heart cell contain the same genome of 46 chromosomes but are likely to differ in their patterns of epigenetic modifications
E) Epigenetic changes are permanent