Lecture 10 - Phylogenies (continued)
1/7
Earn XP
Description and Tags
included on exam 2
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
8 Terms
how to form maximum likelihood values (with bootstrapping)
when several trees are made via bootstrapping, count how many times a certain relationship appears
turn that number into a decimal, then multiply by 100 to get a percentage without the % sign
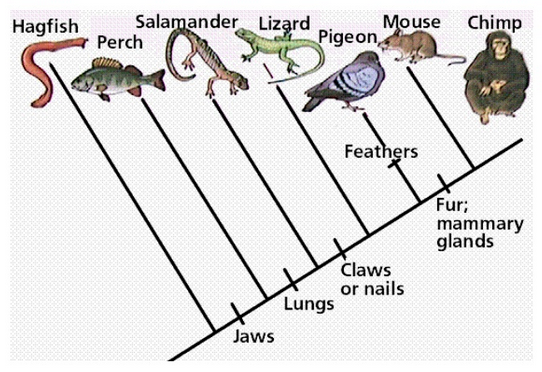
cladogram
any type of phylogenetic tree where the tips all line up with each other
branched lengths are irrelevant, don’t convey any information
phenogram
a phylogenetic tree where the tips don’t all align
still shows the relationships between species, but also tells more information
branch lengths are informative, typically the number of nucleotide differences between species
i.e. shorter branches have fewer base pair differences than long branches
(note: branch lengths are in comparison to each other, not just to the MRCA)
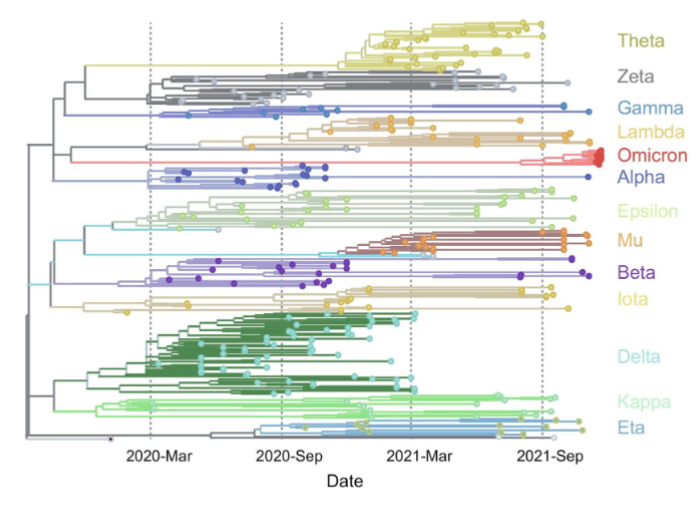
ultrametric tree
a phylogenetic tree that dates a phenogram
uses a molecular clock to calibrate the dating (e.g. establish that 1 nucleotide change happens every 1 million years)
tips of extant taxa are aligned, but not extinct species
consensus
a tree that summarizes a collection of data or other phylogenetic trees
generally not illustrated on node supports
how genetic trees and species trees may differ
gene duplication
gene loss
horizontal gene transfer
how to date a phylogenetic tree
calibrate the molecular clock with: fossils, geographical activity (e.g. volcanic activity or island formation), real-time data
(note: fossils are the most common method)
a fossil can be found that looks like an ancestor to a species
we know the age of the fossil with uniform distribution or log normal
the speciation event can be placed based on that fossil
log normal
a method of dating a phylogeny based on a fossil
a time window is given for the fossil’s age, with a definite youngest age and a most likely older age
but it is possible that the fossil could be older than that window, with decreasing probability
better than uniform distribution