D1.1 DNA Replication | IB Biology HL
1/26
Earn XP
Description and Tags
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
27 Terms
State that DNA replication is the production of exact copies of DNA with identical sequences
D1.1.1: DNA replication as a production of exact copies of DNA with identical base sequences
DNA replication forms exact copies of DNA with identical sequences.
Outline the purposes of DNA replication.
D1.1.1: DNA replication as a production of exact copies of DNA with identical base sequences
DNA replication produces two identical DNA molecules from one original DNA molecule.
DNA replication helps in the inheritance process by transferring genetic material from one generation to another. It is required for the growth, repair, and generation of tissues in living organisms.
Describe the meaning of “semi conservative” in relation to DNA replication.
D1.1.2: Semi-conservative nature of DNA replication and role of complementary base pairing
Semi-conservative replication is the method of DNA replication in which parental strands separate and act as templates. Thus, they produce molecules of DNA with one parental DNA strand and one new DNA strand.
Explain the role of complementary base pairing in DNA replication
D1.1.2: Semi-conservative nature of DNA replication and role of complementary base pairing
The enzyme DNA Polymerase III builds the new strand by “reading” the template & adding the complementary DNA nucleotide. Therefore, the newly built strand will have the same sequence of bases as the other template strand.
State why DNA strands must be separated prior to replication.
D1.1.3: Role of helicase and DNA polymerase in DNA replication
The separation of DNA strands allows DNA to be exposed for base-pairing, allowing DNA polymerase III to build new nucleotides and replication to occur.
Outline the role of helicase in DNA replication.
D1.1.3: Role of helicase and DNA polymerase in DNA replication
Helicase “unzips” the DNA by breaking the hydrogen bonds between base pairs at specific regions (origins of replication), creating a replication fork of two strands running in antiparallel directions.
Helicase moves in both directions in a replication bubble
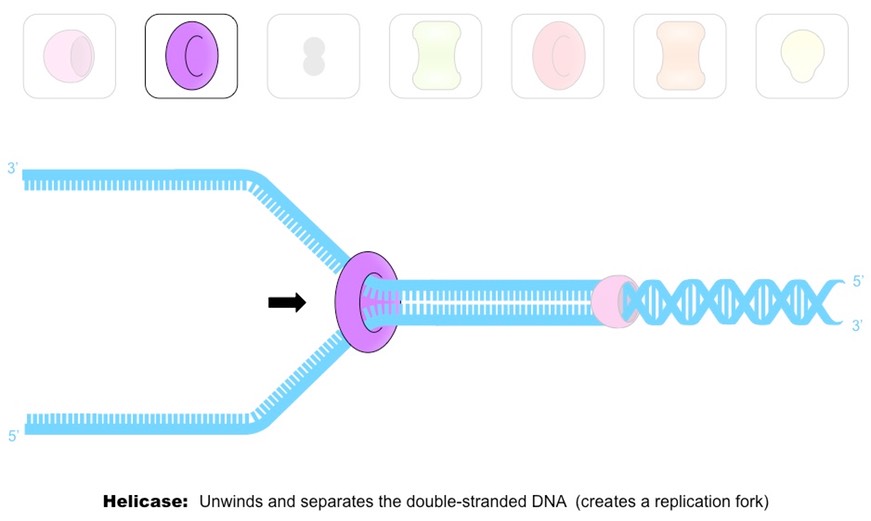
Outline the role of DNA polymerases in DNA replication.
D1.1.3: Role of helicase and DNA polymerase in DNA replication
DNA Polymerase III attaches to the 3’-end of the primer and covalently joins the free nucleotides together in a 5’ → 3’ direction
DNA Polymerase I removes the RNA primers from the lagging strand and replaces them with DNA nucleotides.
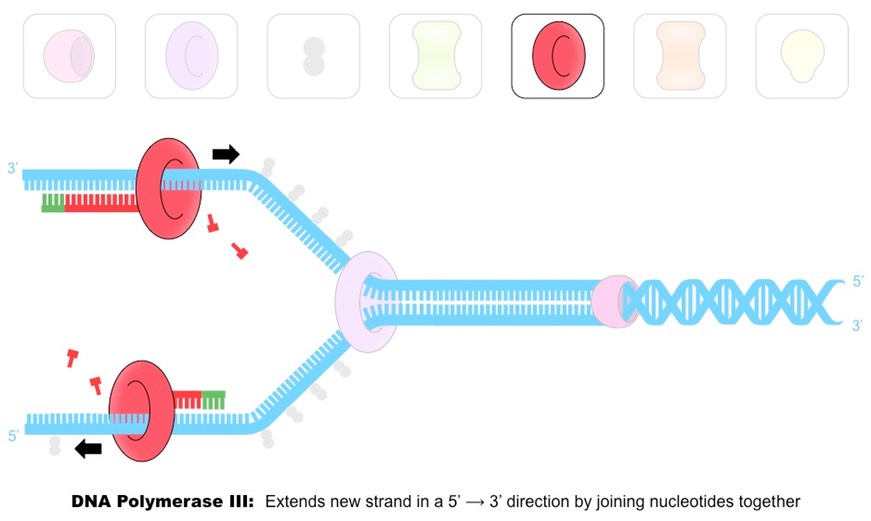
State that there are separate DNA polymerases for each strand of template DNA.
D1.1.3: Role of helicase and DNA polymerase in DNA replication
There are separate DNA polymerases for each strand of template DNA.
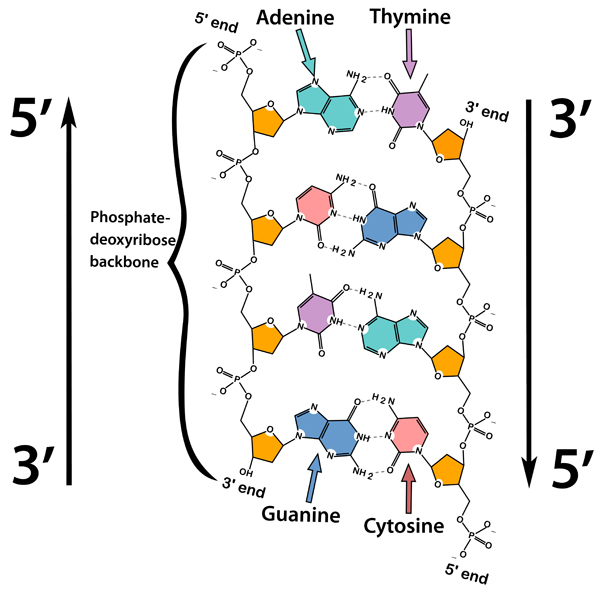
Identify the 5’ ends and 3’ ends of a strand of DNA.
D1.1.6: Directionality of DNA Polymerases
The 5’ end of DNA has the phosphate groups, while the 3’ end is the pentose sugar.
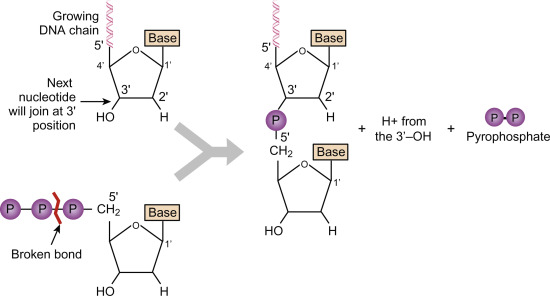
Describe the formation of the covalent bond between adjacent nucleotides during DNA replication
D1.1.6: Directionality of DNA Polymerases
Deoxynucleoside Triphosphate (dNTP) are like a DNA nucleotide, but with three phosphate groups instead of one. They freely float in the nucleus, but when a nucleotide is needed, 2 phosphate groups get broken off. This releases the energy required to form the covalent bond between adjacent nucleotides.
State that DNA polymerases can only add free nucleotides to an existing DNA strand.
D1.1.6: Directionality of DNA Polymerases
DNA polymerases can only add free nucleotides to an existing strand.
This is why DNA primase needs to create an RNA primer first.
State that DNA polymerases can only add the 5’ phosphate of a free nucleotide to the 3’ deoxyribose of the elongating strand.
D1.1.6: Directionality of DNA Polymerases
DNA polymerases can only add the 5’ phosphate of a free nucleotide to the 3’ deoxyribose of the elongating strand.
Explain why replication is different on the leading and lagging strands of DNA.
D1.1.7: Differences between replication on the leading strand and the lagging strand
DNA is synthesized continuously on the leading strand because DNA Polymerase III is moving in the same direction as the replication fork is opening.
DNA is synthesized in pieces (Okazaki fragments) on the lagging strand because DNA Polymerase III is moving in the opposite direction as the replication fork is opening, so polymerase is always catching up to the fork.
Compare the pace and direction of replication on the leading and lagging strands of DNA.
D1.1.7: Differences between replication on the leading strand and the lagging strand
Both the leading and lagging strands are replicated in the 5’ to 3’ direction.
Outline the formation of Okazaki fragments on the lagging strand.
D1.1.7: Differences between replication on the leading strand and the lagging strand
Okazaki fragments are the short fragments that the lagging strand is copied in because polymerase III only replicates DNA in the 5’ to 3’ direction. On the lagging strand, this is opposite to the way helicase is unzipping the DNA. Therefore, DNA primase has to keep putting down primers as DNA is opening, and DNA Polymerase has to keep adding nucleotides in fragments.
Explain the need for RNA primers in DNA replication.
D1.1.8: Functions of DNA primase, DNA polymerase I, DNA polymerase III and DNA ligase in replication.
RNA primers provide an initiation point for DNA polymerase III, which can extend a nucleotide chain but cannot start one. Polymerase III attaches to the 3’-end of the primer and attaches new DNA nucleotides to the primer.
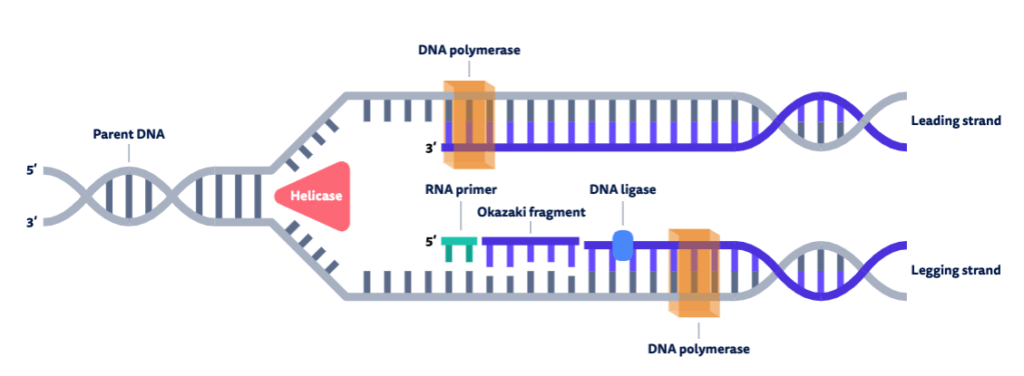
Compare the number of RNA primers on the leading and lagging strands.
D1.1.8: Functions of DNA primase, DNA polymerase I, DNA polymerase III and DNA ligase in replication.
The leading strand has one RNA primer, while the lagging strand has multiple.
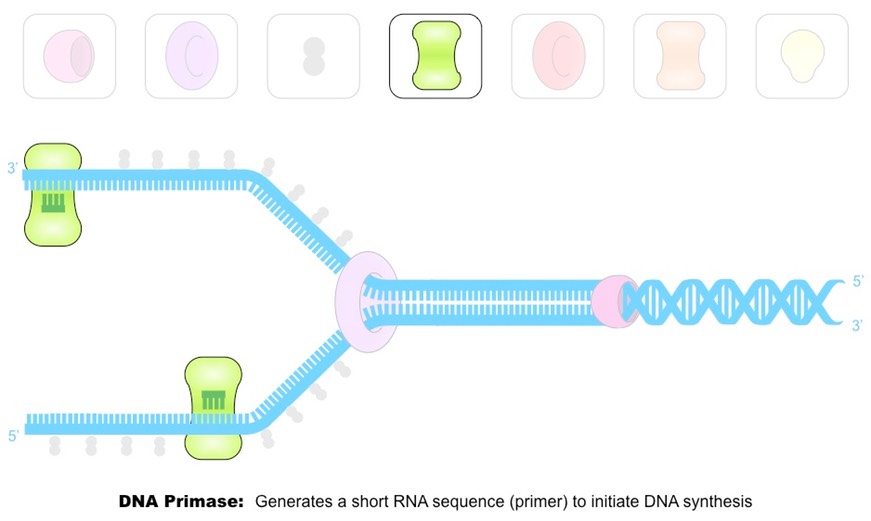
Outline the function of the enzyme DNA primase.
D1.1.8: Functions of DNA primase, DNA polymerase I, DNA polymerase III and DNA ligase in replication.
DNA Primase generates a short RNA primer (about 10-15 nucleotides) on each of the template strands. The RNA primer provides an initiation point for DNA polymerase III, which can extend a nucleotide chain but not start one.
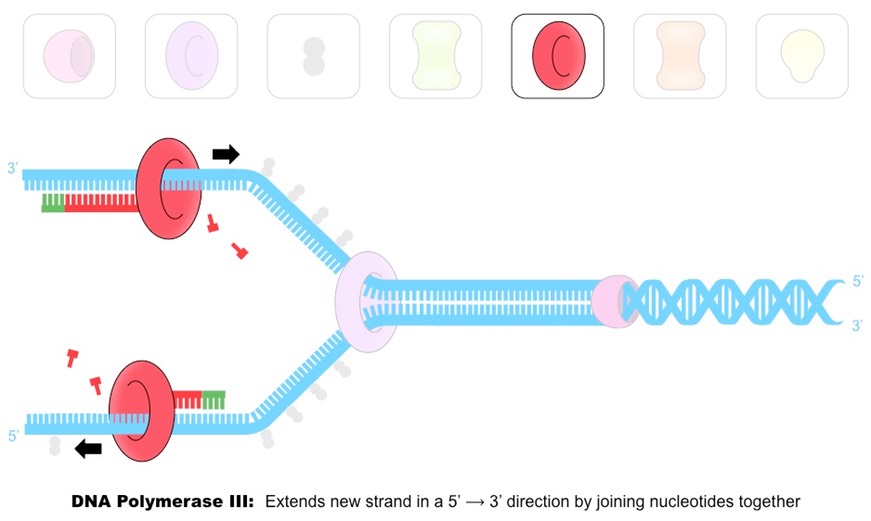
Outline the function of the enzyme DNA polymerase III.
D1.1.8: Functions of DNA primase, DNA polymerase I, DNA polymerase III and DNA ligase in replication.
DNA polymerase III attaches to the 3’ end of the RNA primer and covalently joins free complementary base nucleotides in a 5’ to 3’ direction with phosphodiester bonds.
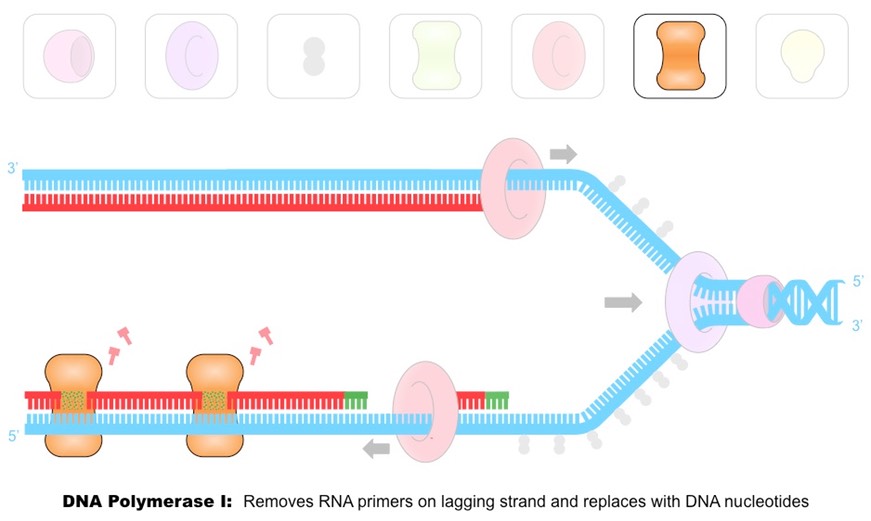
Outline the function of the enzyme DNA polymerase I.
D1.1.8: Functions of DNA primase, DNA polymerase I, DNA polymerase III and DNA ligase in replication.
DNA polymerase I removes the RNA primers and replaces them with DNA nucleotides.
Explain why there are gaps between adjacent Okazaki fragments on the lagging strand.
D1.1.8: Functions of DNA primase, DNA polymerase I, DNA polymerase III and DNA ligase in replication.
The ends of Okazaki fragments do not form covalent bonds with the RNA primer of the next Okazaki fragment, forming gaps between the fragments.
Outline the function of the enzyme DNA ligase.
D1.1.8: Functions of DNA primase, DNA polymerase I, DNA polymerase III and DNA ligase in replication.
DNA Ligase joins Okazaki fragments and the new DNA nucleotides (which replaced the RNA primers) to the rest of the strand with a phosphodiester bond (to join the backbones together) to form a continuous strand of DNA.
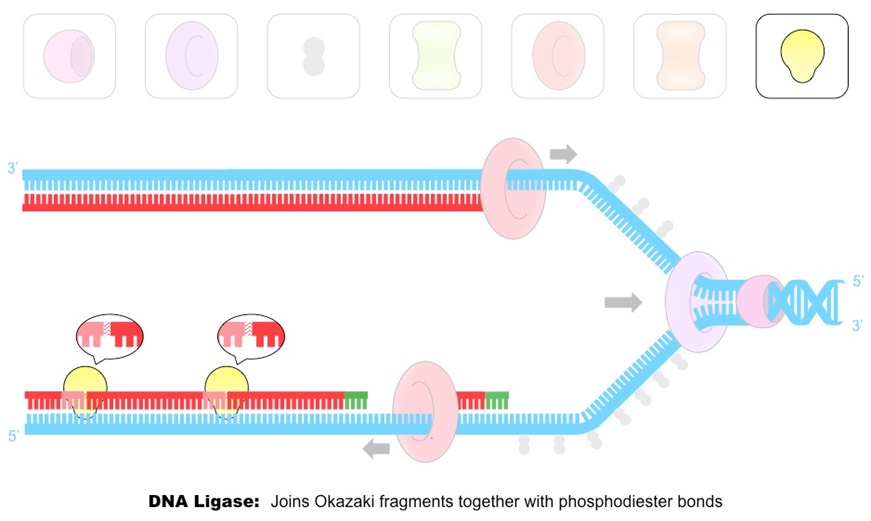
Outline the steps of DNA replication.
DNA replication
Helicase unzips the DNA by breaking bonds between nitrogen bases, creating a replication fork.
DNA Gyrase unwinds DNA to release tension.
Single Stranded Binding (SSB) proteins bind to DNA strands to keep strands from winding back together and to protect DNA from chemical damage.
DNA Primase generates a short RNA primer (as an initiation point for Polymerase III)
DNA Polymerase III attaches to the 3’ end of the primer and adds new DNA complementary base pairs.
DNA Polymerase I remove the RNA primers from the lagging strand and replaces them with DNA nucleotides.
DNA Ligase joins the Okazaki fragments and fragments of DNA (that replaced RNA primers) with phosphodiester bonds to form a continuous strand.
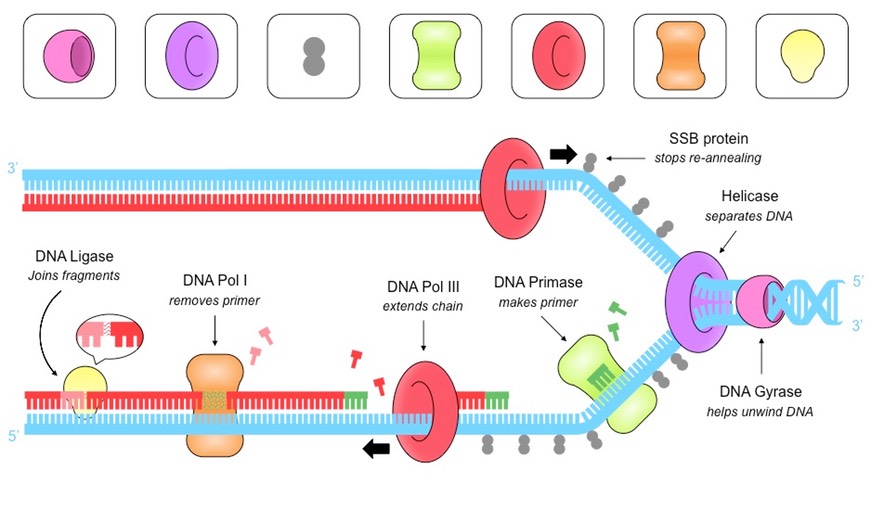
List the enzymes involved in DNA replication and their functions.
DNA Replication
Helicase: Unwinds the double helix at replication forks
DNA Gyrase: Reduces tension by unwinding DNA before Helicase.
Single Stranded Binding (SSB) Proteins: Binds to DNA strands after they’ve been separated to prevent them from winding back together and prevent chemical damage.
DNA Primase: Generates a short RNA primer (to provide an initiation point for DNA Polymerase III)
DNA Polymerase III: Attaches to 3’-end of the primer and covalently joins free nucleotides (with the complementary base pairs) in a 5’ to 3’ direction
DNA Polymerase I: Removes the RNA primers form the lagging strand and replaces them with DNA nucleotides.
DNA Ligase: Joins Okazaki fragments and new DNA strands with a phosphodiester bond to form a continuous strands (like glue)
How do cells ensure accuracy in replication?
DNA Replication
Cells have a number of repair enzymes that detect and correct errors when they do occur.
These repair enzymes are also used when chemicals or high-energy waves cause damage to existing cells.
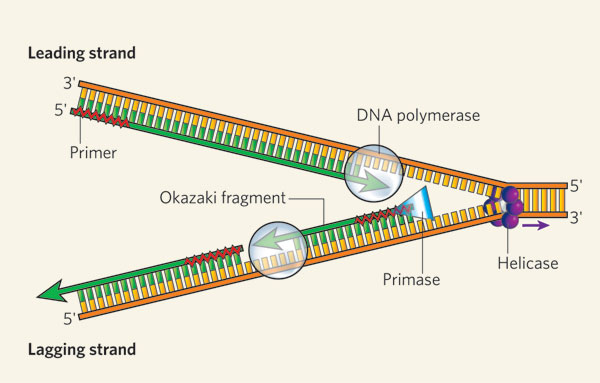
Distinguish between the leading and lagging strand.
DNA Replication
Leading strand: opens in the 5’ to 3’ direction towards the replication fork, synthesized continuously
Lagging strand: opens in the 3’ to 5’ direction towards the replication fork, is synthesized discontinously in Okazaki fragments
Outline what provides the energy for the chemical bonding of nucleotides.
DNA Replication
The two extra phosphates attached to the deoxynucleoside triphosphate (dNTP) molecule release energy when they’re broken off to combine nucleotides.