LF206 lecture 1 - bacterial genomes
1/27
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
28 Terms
the traditional view of prokaryotic genome (based on ______) was a single, c______ DNA molecule localised within the _________.
E.coli
circular
nucleoid
a majority of ___ in living cells occurs in the B form.
what is it?
DNA
2 polynucleotide chains in opposite orientation
regular right-handed double helix
diameter of 2nm and makes a complete turn every 3.4 nm
~10.5 base pairs per turn of the helix
there are certain flexibilities within the basic B-form
the number of base pairs ___ ____ of the helix can be altered
the helix in the cell is not _______ but curled in __ space
per turn
straight
3D
DNA in the ______ genome of E. coli is supercoiled
_______ supercoiling occurs when additional turns are introduced into the DNA double helix when twisted in a ____-handed fashion.
_______ supercoiling occurs when turns are _______ and are twisted in a left-handed fashion
circular
positive, right
negative, removed
define torsional stress
when a twisting force (torque) is applied causing internal stress to resist the twisting
when DNA undergoes torsional stress how is it accommodated? [2]
formation of superhelices (a helix coiling itself into a helix)
altering the number of base pairs per turn of helix
define linking number
total number of times that both strands of the double helix (of a closed molecule) cross each other when constrained to lie in a plane
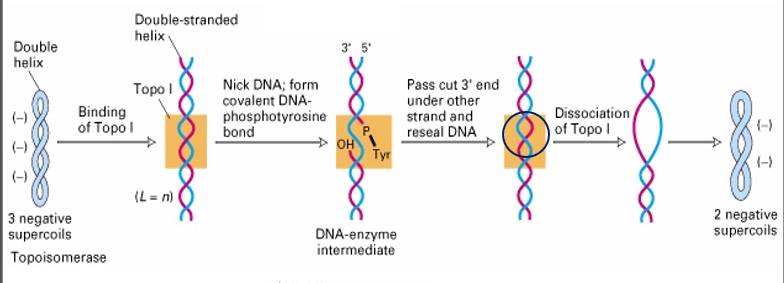
Type I Topoisomerases break ___ strand of DNA and create a covalent DNA-____________ bond.
then pass the _ ‘strand under the other strand and ______ the _____.
the ______ number is changed by +or- 1.
Type _ topoisomerase of E.coli _______ly supercoils DNA
one
phosphotyrosine
3
seal
break
linking
I
negatively
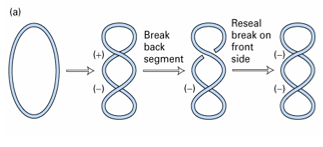
Type __ topoisomerases break both strands of ___ and pass _______ ___ of the helix through the gap. the linking number changes by + or - ___.
DNA ______ of E.coli is a hetero________ of 2 subunits (which are?) creates negative supercoils using ___
II
DNA
another part
2
gyrase
heterotetramer
A and B
ATP
topoisomerases are _______ that alter the ______ (shape) of DNA .
they often relieve _______ stress by creating transient (define this) breaks in DNA = ______ supercoiling.
but can also introduce positive supercoiling
enzymes
topology
torsional
transient = temporary
negative
how is DNA organised in bacteria (E.coli)?
single circular DNA molecule in a series of supercoiled loops (40-50) radiating from a central protein core
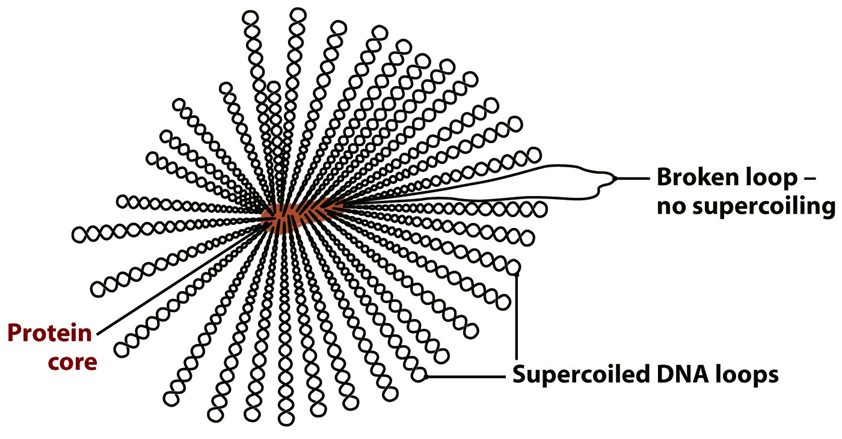
the protein component of the E.coli nucleoid includes
DNA ______ and DNA topoisomerase _ to maintain the ______ state
at least 4 proteins involved in packaging DNA - the most abundant is ______ (HU)
HU forms ______ around what DNA is wound
approximately 1 HU for every __ bp, cover ___ of the genome
______ don’t have protein related to HU, instead they have proteins related to eukaryotic _______
HU is also known as _______-_______ ______ (NAP)
gyrase, I, supercoiled
heat unstable
60 base pairs, 1/5
tetramers
Archaea, histones
nucleoid-associated protein
some bacteria have a linear genome (give 1 example)
some have multipartite genomes (define this)
define plasmid
Borrelia burgdoferi (Lyme disease), Streptomyces coelicolor (antibiotic producer), Agrobacterium tumefaciens (plant tumours)
genomes divided into 2 or more DNA molecules
(often small) DNA molecule that usually codes for non-essential genes - large plasmids can carry essential genes
what is a prophage?
genome of a bacteriophage (a virus that attack bacteria)
what is a replicon?
each replicon must be _________ at least once per cell ______ cycle
the basic unit of replication with a functional origin of replication
replicated
division
many important genes in bacteria have been acquired by __________ ___ ______ (HGT).
examples of HGT
horizontal gene transfer
prophages
genomic islands
transposable genetic elements
genomic islands
acquired by HGT genomic regions often mutated so masking/destroying their modes of transmission (cannot leave the cell) and confer fitness to occupy a niche.
transposable genetic elements can ____ from one location in the genome to another
‘jump’
Cell cycle in E.coli
________ of replication commits the cell to a subsequent _______.
Cell division cannot occur until the round of ______ associated with a particular initiation has been _________.
E.coli has a single ______ __ _________ (oriC) therefore has a single _____.
E.coli replicates __-directionally leading to a ___ structure
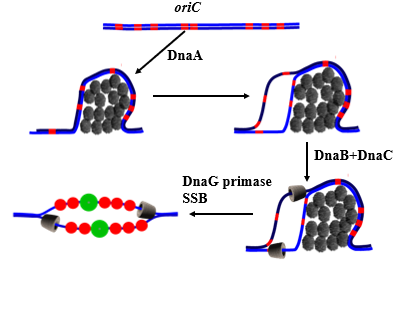
initiation commences with the binding of ~__ monomers of Dna_ to 4 out of the _ 9 base pair repeats in the _______ ______ (RH) part of ____
this forms a _______ complex
an ____ complex forms when the 3 __-rich 13-bp repeats ___
DnaB _______ is loaded onto the melted DNA with _____
ATP is __________ and DnaC is _______
DnaB unwinds DNA ________lly in a process that requires ______ _______ _______ (SSB) and DNA gyrase
______ synthesises a primer RNA molecule on both strands and replication commences
initiation, division
replication, completed
origin of replication, replicon
bi, theta
20, DnaA, 5, ???????, oriC
closed
open, AT, melt
helicase, DnaC
hydrolysed, released
bidirectionally, Single Strand Binding protein
primase
what controls whether a round of replication is initiated?
replication only continues if all 14 copied of ____ in oriC are __________.
____ _________, methylates adenine residues GATC motifs in oriC.
after replication, GATC copies on the _____ ___________ strand are not methylated.
DNA molecules are ____-methylated as the old strand is methylated and the new strand is ___________. this is governed by semi-_________ replication.
hemi-methylated oriC DNA is sequestered at a ________site and is not available for replication
re-methylation occurs ~_____ of the way into the cell cycle
GATC
methylated
Dam methylase
newly synthesised
hemi-methylated
unmethylated
semi-conservative
membrane
1/3
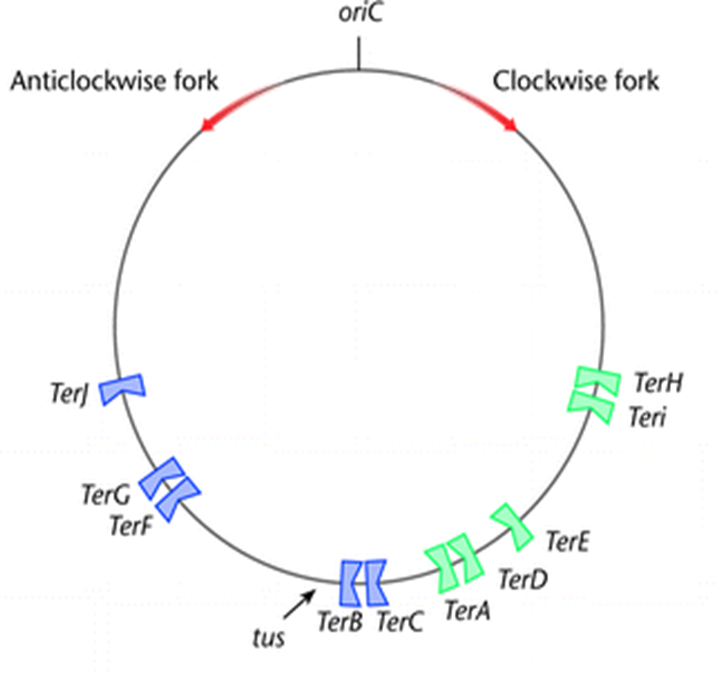
how is replication terminated?
both replication forks (which began at oriC) approach one another and fuse at the terminus region.
terminus region is a replication fork trap
has a series of DNA sites that arrest fork progression
DNA ___________ (ter) sites are polar in their action - meaning what?
___, a terminator protein, must be bound to the ter site to ____ ___ _____.
In E.coli the ter sites are distributed over ____% of the chromosome but in B. subtilis they are spread much ________er (what percentage?)
terminator
they will arrest a fork approaching from one direction but not the other
Tus
halt the fork
42.5%
narrower
9.9%
this E.coli DNA,
what sites does the clockwise fork pass?
what sites does the clockwise fork halt? - if it doesn’t initially halt there, where else will it?
terH, i, E,D and A
terC + Tus
should halt when anticlockwise fork arrives but will continue to terB
if anticlockwise doesn’t arrive, clockwise continues after a pause and terJ is the last opportunity to terminate.
why is a fork arrested from only 1 direction?
Tus protein can only inhibit helicase (at the apex of the fork) in one direction.
inhibiting helicase prevents DNA from unwinding and therefore replicating.
_____________ recombination is essential for the generation of _______ diversity and ___ repair
for recombination to occur the 2 molecules must have ________ ______ of the order (approx.) of 100-500bp
homologous
genetic
DNA
homologous regions
Meselson-Radding model
one DNA strand is cleaved by an endonuclease
DNA synthesis displaces a chain
the single stranded chain invades a homologous double-stranded DNA molecule - catalysed by RecA
the displaced chain is digested
remaining single strand gaps are ligated producing a Holliday junction
branch migration catalysed by RuvAB increases DNA heteroduplex
isomerization: strands of the Holliday junction spontaneously cross and uncross - no catalysis required.
resolution: crossed strands of Holliday junction are cleaved by RuvC
unlike Holliday model, products can be asymmetrical
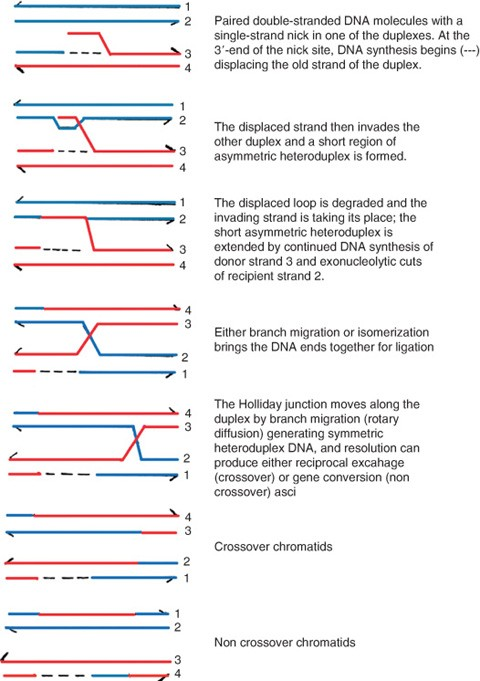
DOUBLE STRAND HOMOLOGOUS RECOMBINATION
RecBCD binds to the end of a ______ substrate and unwinds using its ______ activity
as RecBCD unwinds the helix, it ______ both ______ strands using its dual _’→_’ and _’→_’ ___nuclease activities
RecBCD moves in _____ and each ____ is __ bp long. this is called the ________ _______
when RecBCD encounters a χ (what symbol is this and what is it?) site its enzymatic activities are altered
_’→_’ exonuclease activity is inhibited
5’ → 3’ exonuclease activity is _______
helicase activity is _______
RecBCD produces a ssDNA___ with a _’ end and RecA binds to the ssDNA tail which ______
dsDNA, helicase
degrades, ssDNA, 3’→5’, 5’→3’, exonuclease
steps, step, 23, quantum inchworm
chi, sites of recombination 5’ GCTGGTGG 3’ (there are 1009 chi sites on the E.coli genome)
3’→5’
stimulated
unaffected
tail, 3’, invades
DNA sequences which do not transpose alter their orientation within a ___ molecule - __________ sequences.
most Salmonella spp. can produce two different types of ________
phase _ and _ _ antigens
phase is determined by the orientation of ___ (H-inversion) region
this region is ___ bp long and bounded by two ___ bp _______ repeats
this region encodes for _______ which catalyses inversion
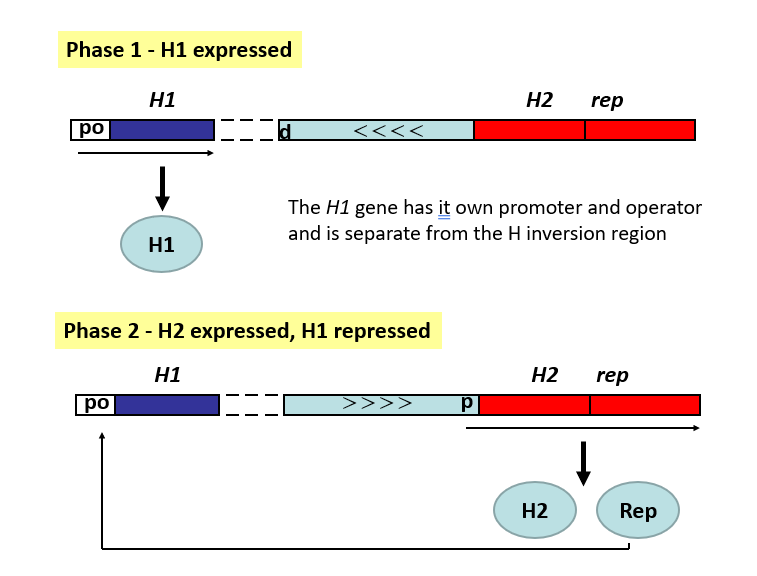
differences between H1 and H2 genes
DNA
inversion
flagellum
phase 1 and 2 H antigens
hin
992
14
inverted
invertase
H1 gene has its own promoter and operator & is physically separated from the hin region
H2 gene is in an operon with the rep gene that encodes a repressor for H1 gene
H2-rep promoter lies within the hin region