MBS RNA module 3
1/257
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
258 Terms
You treat RNA with DMS, which methylates unpaired adenines (A) and cytosines (C), then pertorm sequencing to map modifications (DMS-seq). A particular loop in your RNA shows low DMS signal. What is the most likely interpretation?
The loop is protected due to protein binding or base-pairing.
You perform an RNase digestion assay using RNase V1 (cuts double-stranded RNA) and RNase 1 (cuts single-stranded RNA) to probe the structure of an RNA molecule. After running a gel, you notice that a region previously predicted to be single-stranded shows strong cleavage with RNase V1 but not RNase 1. What does this result most likely
The region is actually double-stranded, not Single-stranded
A researcher designs an RNA aptamer that binds specifically to a disease-related protein. How could this aptamer be used in drug therapy?
By binding to the target protein and klocking its activity
The same RNA sequence can fold into two different ribozymes with distinct catalytic activities. What does this observation suggest about RNA?
RNA can adopt multiple structures that dictate function
RNA double helices are typically wider and adopt an A-form geometry, unlike the B-form seen in most DNA. What primarily causes this difference?
The 2'-OH on ribose in J RNA causes steric hindrance
DMS-seq and SHAPE-seq can be performed directly in living cells. Why is it beneficial to study RNA structure in vivo rather than only in vitro?
RNA folding cells is influenced by proteins and other molecules
A researcher wants to study the secondary structure of an RNA molecule. She considers three approaches: RNase digestion, DMS-seq, and SHAPE-seq. Which of the following best compares the methods in terms of what they detect and how they provide structural information?
RNase digestion cleaves specific RNA regions;
DMS-seq modifies accessible C and A;
SHAPE-seq modifies 2' OH in ribose.
A molecular biologist treats an RNA molecule with different RNases that selectively cleave either single-stranded or double-stranded RNA. What is the primary purpose of performing RNase digestion in RNA studies?
To map RNA secondary structure
When comparing the stability of different regions in an RNA molecule, a researcher notices that segments rich in guanine (G) and cytosine (C) remain intact at higher temperatures than regions rich in adenine (A) and uracil (U). Which property best explains why G-C base pairs are the strongest in
G-C pairs form more hydrogen bonds than A-U pairs
The RNA 1° structure is held together via?
phosphodiester bonds
A predicted hairpin has a AG of -20 kcal/mol, while another loop has a AG of -5 kcal/mol. What does this indicate about their relative stability?
The hairpin is more stable and more likely to form
Which structural element involves base-pairing between two separate hairpin loops?
Kissing loops
What percentage of total RNA in a cell is translated into protein?
<5% of cellular RNA is translated; most RNA is noncoding.
What is transcription in prokaryotes?
the process in which RNA is synthesized from a DNA template by RNA polymerase. It occurs in the 5′→3′ direction, using the template DNA strand, and does not require a primer.
How does RNA polymerase differ from DNA polymerase?
does not need a primer, uses only one DNA strand as a template, works outside S-phase, and is more error-prone (1 error per 10⁴ bases vs 10⁷ in replication).
What is the typical organization of a prokaryotic operon?
It includes a promoter, structural genes (ORFs or cistrons), and a terminator. The mRNA is often polycistronic, encoding multiple proteins from one transcript.
Define “cistron.”
a coding region or open reading frame (ORF) that corresponds to one gene within an operon.
What enzyme transcribes all bacterial genes?
A single multi-subunit RNA polymerase performs transcription for all genes in bacteria.
What are the subunits of bacterial RNA polymerase?
The core enzyme consists of two α subunits, one β, one β′, and one ω. When combined with a σ factor, it forms the holoenzyme (~450
What is the role of the σ (sigma) factor in transcription?
directs RNA polymerase to specific promoter sequences to initiate transcription. It is released after about 10 nucleotides are added.
How many σ-factors exist in E. coli and why are they important?
has 7 σ-factors, each recognizing distinct promoter sequences for genes expressed under different conditions (e.g., housekeeping, stress response).
How does the number of σ-factors vary among bacterial species?
It varies widely—Mycoplasma genitalium has 1, Bacillus subtilis has 13, and Streptomyces coelicolor has over 60.
What are the three main stages of transcription?
Initiation, Elongation, and Termination.
What occurs during transcription initiation?
RNA polymerase binds the promoter (closed complex), unwinds DNA to form a transcription bubble (open complex), forms the first phosphodiester bonds, and releases the σ factor to form a stable ternary complex.
What is the function of the promoter?
The promoter is a DNA sequence upstream of a gene that signals RNA polymerase where to begin transcription.
What are the typical bacterial promoter regions recognized by σ⁷⁰
Two conserved sequences: the −35 region (TTGACA) and the −10 region (Pribnow box: TATAAT).
What is the UP element and its function?
An A+T-rich region (~20 bp) located upstream of −35 that binds the α subunit’s C-terminal domain, strengthening promoter binding in highly expressed genes (e.g., rRNA genes).
What is abortive initiation?
Early rounds of transcription that produce short (~10 nt) RNA fragments due to δ-factor blocking the RNA exit channel. Productive transcription begins only after promoter escape.
What is promoter escape?
The transition from abortive to productive transcription when RNA polymerase clears the promoter and forms a stable ternary complex (DNA-RNA-RNAP).
What direction does RNA chain elongation occur in?
5′ → 3′ direction, adding nucleotides to the 3′-OH end of the growing RNA strand
What is the average elongation rate of RNA polymerase in E. coli?
40–80 nucleotides per second in vivo.
What happens to DNA topology during transcription
Positive supercoils form ahead of the transcription bubble, and negative supercoils form behind it. Topoisomerases resolve these tensions
How processive is RNA polymerase during elongation?
It remains tightly bound to DNA until termination, forming a highly stable ternary complex with ~14 nucleotides of RNA protected within RNAP.
Why is elongation not a smooth process?
RNA polymerase often pauses due to RNA secondary structures, difficult sequences, backtracking, or limited NTP concentrations.
Does RNA polymerase have proofreading ability?
Yes, but it’s limited—transcription is more error-prone than DNA replication.
How is transcription coupled to translation in bacteria?
Ribosomes begin translating the nascent mRNA while it is still being transcribed, allowing simultaneous transcription and translation.
What signals transcription termination?
Specific DNA sequences called terminators that cause RNA polymerase to stop and release the transcript.
What are the two types of transcription termination in bacteria?
Intrinsic (rho-independent) and rho-dependent termination.
Describe intrinsic (rho-independent) termination.
A GC-rich inverted repeat followed by a string of A’s forms a hairpin loop in the RNA, stalling RNAP and weakening the AU-rich RNA–DNA hybrid, leading to transcript release.
What stabilizes and destabilizes intrinsic terminators?
The GC-rich hairpin stabilizes the RNA structure; the AU base pairs downstream are weak, facilitating RNA release.
What enzyme mediates rho-dependent termination
The Rho factor, an ATP-dependent helicase that unwinds RNA–DNA hybrids and releases the transcript.
What is the rho utilization (rut) site?
A C-rich, ~40-nt sequence on nascent RNA where Rho binds to initiate termination.
How does rho terminate transcription?
Rho moves 5′→3′ using ATP, catches up with the stalled RNA polymerase, unwinds the RNA–DNA hybrid, and releases the RNA.
What causes RNA polymerase stalling that allows Rho to catch up?
Hairpin formation or GC-rich sequences that slow polymerase movement.
What happens after transcription termination?
The RNA polymerase detaches from DNA, and RNA may undergo limited post-transcriptional processing.
How are rRNAs processed in bacteria?
All rRNAs are derived from a single precursor molecule that undergoes cleavage and base modifications to yield mature rRNAs.
Why is post-transcriptional mRNA processing rare in bacteria
Because transcription and translation are coupled, leaving little time for processing. However, some modifications like polyadenylation do occur.
What is the role of polyadenylation in bacteria?
Unlike in eukaryotes, bacterial polyA tails promote RNA degradation rather than stability.
What are the main enzymes involved in RNA degradation?
Endonucleases (RNAse E, RNAse III)
and
exonucleases (RNAse II, PNPase).
What is the RNA degradosome?
A multiprotein complex consisting of RNases and RNA helicase that coordinates RNA degradation in bacteria
Why are bacterial mRNAs short-lived
Their half-lives are only a few minutes, ensuring rapid turnover and adaptability to environmental changes.
How does transcription efficiency vary among bacterial genes?
Different genes are transcribed at varying times and rates, regulated by promoter strength and σ-factor availability.
Summarize the key differences between transcription and replication
Transcription uses RNA polymerase (no primer, one DNA strand, 5′→3′), is continuous, error-prone, and coupled to translation.
Replication uses DNA polymerase, requires a primer, copies both strands, and is cell-cycle restricted.
Why do bacteria regulate gene expression?
To conserve resources and respond to environmental changes; not all 4200 E. coli genes are expressed all the time.
Define constitutive, inducible, and repressible expression.
Constitutive: Always on
Inducible: Normally off; can be turned on by an inducer
Repressible: Normally on; can be turned off by a repressor
Main levels of regulation in bacteria?
Transcription regulation
mRNA stability
Translation efficiency
What are the structural genes of the lac operon?
lacZ, lacY, and lacA, coding for β-galactosidase, permease, and transacetylase.
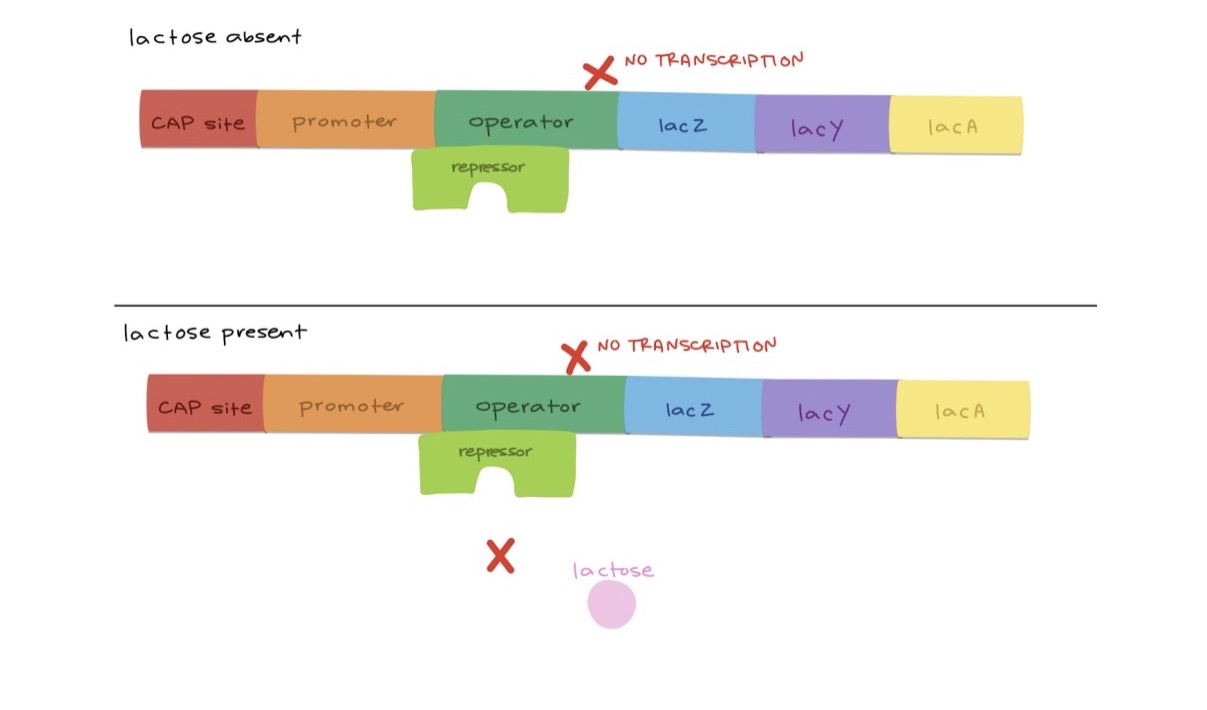
What happens to the lac operon when lactose is absent?
The lac repressor binds to the operator, preventing RNA polymerase from binding the promoter — transcription is inhibited.
What happens when lactose is present?
Allolactose (derived from lactose) binds the repressor, changing its shape so it cannot bind the operator — RNA polymerase binds the promoter and transcribes lac genes.
How does glucose affect lac operon expression?
High glucose → low cAMP → CAP cannot bind → weak RNA Pol binding → low lac transcription
Low glucose → high cAMP → CAP-cAMP binds → strong RNA Pol binding → high lac transcription
What is CAP and its role?
Catabolite Activator Protein; binds to the CAP site when complexed with cAMP to enhance RNA polymerase binding and increase transcription.
What are the key genes in the trp operon?
Five structural genes that encode enzymes for tryptophan synthesis, plus a leader sequence and an operator region.
Trp Operon (Repressible System)
What happens when tryptophan levels are low?
Repressor cannot bind the operator → RNA polymerase transcribes trp genes → tryptophan synthesized.
Trp Operon (Repressible System)
What happens when tryptophan levels are high?
Tryptophan binds the repressor → repressor changes conformation → binds operator → transcription blocked.
Trp Operon (Repressible System)
What is transcriptional attenuation?
A secondary regulatory mechanism in which transcription terminates early depending on translation of a leader peptide that includes two Trp codons.
Trp Operon (Repressible System)
What happens to attenuation when tryptophan is low?
Ribosome stalls at Trp codons → alternative (non-terminator) loop forms → transcription continues.
Trp Operon (Repressible System)
What happens to attenuation when tryptophan is high?
Ribosome does not stall → terminator hairpin forms → transcription stops.
What are small RNAs (sRNAs)?
Short (~80–100 nt) noncoding RNAs that base-pair (imperfectly) with target mRNAs to regulate translation or stability, often assisted by Hfq chaperone.
All three methods-RT-PCR, 9RT-PCR, and RNA-seq —share which of the following properties?
They involve cDNA synthesis from RNA
In gRT-PCR, a sample shows a Ct value of 18 cycles, while another shows a Ct of 28 cycles for the same gene. What does this suggest?
The sample with Ct = 18 has more starting RNA
What base pairs is not seen in RNA molecules?
Adenine–Thymine (A–T)
A researcher is studying an RNA molecule that folds into a complex 3D structure, forming
loops, stems, and bulges. When the RNA is denatured and loses its shape, its catalytic activity disappears. What does this observation suggest about the importance of RNA’s ability to adopt different shapes?
The folded shapes of RNA enable it to perform diverse functions
tRNA molecules often contain unusual or modified bases, such as inosine, pseudouridine, or methylated nucleotides. Why are these modified bases most commonly found in tRNA?
They stabilize tRNA’s 3D structure and improve accurate codon recognition
A molecular biologist treats an RNA molecule with different RNases that selectively cleave either single-stranded or double-stranded RNA. What is the primary purpose of performing Rnase digestion in RNA studies?
To map RNA secondary structure
: RNase digestion is commonly used to analyze RNA structure. By using
RNases with different specificities (e.g., single-strand–specific RNase A or double-strand–specific RNase V1), researchers can determine which regions of an RNA molecule are single-stranded versus double-stranded, providing insight into its secondary and tertiary structures.
What is the main role of RNA molecules in the cell?
Translation
RNA molecules, especially mRNA, tRNA, and rRNA, play a central role in
translation, the process of synthesizing proteins from genetic information. While RNA is
involved in transcription and can influence gene modification, its primary functional role
is carrying and interpreting genetic information to produce proteins.
What is the primary function of a hammerhead ribozyme?
Cleaving RNA at specific sites without protein enzymes
These ribozymes are small self-cleaving RNA molecules that can cut RNA at specific sequences without the need for protein enzymes.
A researcher designs an RNA aptamer that binds specifically to a disease-related protein.
How could this aptamer be used in drug therapy?
By binding to the target protein and blocking its activity
these are short RNA molecules that bind tightly and specifically to
target molecules. In therapy, they can inhibit disease-related proteins, similar to how
antibodies work, making them useful as targeted drugs or molecular inhibitors.
.A researcher wants to study gene expression and is deciding between RT-PCR and qRT-PCR. What is the main benefit of using qRT-PCR compared to conventional RT-PCR?
qRT-PCR can measure the amount of RNA in real time
Both RT-PCR and qRT-PCR start by converting RNA to cDNA using reverse transcriptase. The key advantage of qRT-PCR (quantitative or real-time PCR) is that it monitors the amplification in real time, allowing researchers to quantify RNA levels, whereas conventional RT-PCR only detects presence or absence after the reaction is complete
In a qRT-PCR experiment, a researcher observes that a sample reaches the fluorescence threshold at a low Ct value. What does this indicate about the RNA in that sample?
The RNA was present at a high initial concentration
A low Ct value means the target RNA was ?
abundant at the start, because fewer cycles were needed to reach detectable levels.
.A researcher uses RNA-seq to compare cells that are treated with a drug versus untreated cells. Why is RNA-seq useful for this ?
It shows which genes are turned on or off and how much they are expressed
It measures the amount of every RNA transcript in a sample, so researchers can see which genes are upregulated or downregulated in treated versus untreated cells.
.You treat RNA with DMS, which methylates unpaired adenines (A) and cytosines (C), then perform sequencing to map modifications (DMS-seq). A particular loop in your RNA shows low DMS signal. What is the most likely interpretation?
The loop is protected due to protein binding or base-pairing.
It reacts with unpaired A and C bases. When this is low reactivity in a region that should be single-stranded indicates either protection from protein binding or unexpected base-pairing, suggesting structural stabilization.
SHAPE-seq reacts with:
All nucleotides
A bacterial cell is exposed to a sugar. Within minutes, several enzymes needed to metabolize that sugar are produced, all from genes located next to each other on the chromosome. Which of the following best explains how the cell was able to coordinate this response so quickly?
All the enzyme genes were part of a single operon controlled by one promoter and one regulatory region.
In bacteria, this is a group of genes that share a single promoter and regulatory region, allowing the cell to rapidly turn on or off all the genes in a pathway at once.
In bacteria, what is the main role of a sigma factor during transcription initiation?
It helps RNA polymerase recognize and bind to the correct promoter.
This guides RNA polymerase to specific promoter sequences, ensuring the correct genes are transcribed. Once transcription begins, the sigma factor often detaches
Sigma factor
During the early stages of transcription in bacteria, the DNA-RNA polymerase complex is unstable as the first few phosphodiester bonds are formed. Why does this instability
occur?
The sigma factor weakens the interaction between RNA polymerase and DNA until the transcript becomes long enough
Sigma factors help RNA polymerase bind to the promoter, but they also make the initial complex less stable. This allows the polymerase to repeatedly start short transcripts (abortive initiation) until the RNA chain is long enough (~10 nucleotides) to stabilize the complex and allow sigma to release. After sigma dissociates, elongation proceeds
smoothly.
A sigma factor recognizes a specific consensus promoter sequence in bacteria. Three promoters are compared based on how closely they match this consensus: Promoter X: 99% match;
Promoter Y: 89% match;
Promoter Z: 50% match
Which promoter will RNA polymerase bind to most tightly ?
Promoter X
The closer a promoter sequence matches the sigma factor’s consensus sequence, the more strongly RNA polymerase (with its sigma factor) binds to it. A 99% match means This provides the strongest and most stable binding, leading to higher levels of transcription.
Why is it beneficial for bacteria to have multiple sigma factors that recognize different promoter sequences?
It lets bacteria quickly switch on specific sets of genes in response to changing conditions.
Different sigma factors guide RNA polymerase to different promoter sequences. This allows bacteria to rapidly activate the right genes when facing new environments
In bacterial promoters, the –10 region is typically rich in adenine (A) and thymine (T)
bases. Why is this beneficial for transcription initiation?
A–T base pairs form fewer hydrogen bonds
A–T pairs have only two hydrogen bonds (compared to three in G–C pairs), so A–T–rich regions are easier to separate. This helps RNA polymerase open the DNA double helix at the promoter and begin transcription
A scientist uses DNA footprinting to study how RNA polymerase binds to a promoter. She finds that a short section of the DNA is not cut by DNase when RNA polymerase is present. What does this show?
RNA polymerase protects that part of the DNA because it is bound there.
In this , the area where a protein binds is protected from digestion. This reveals the exact region of the DNA where RNA polymerase attaches to start
transcription.
You want to find out where RNA polymerase binds along the bacterial chromosome. Which experimental methods would be most useful for identifying its binding sites on DNA?
DNA footprinting and ChIP-seq
DNA footprinting can identify the exact nucleotides protected by RNA polymerase on a specific DNA fragment, while ChIP-seq maps where RNA polymerase binds across the entire genome.
Both techniques reveal protein–DNA interaction sites.
During the start of transcription in bacteria, RNA polymerase often makes and releases short RNA fragments before making a full transcript. Why does this abortive initiation
happen?
The sigma factor makes the complex unstable.
In early transcription, the sigma factor causes the RNA polymerase–DNA complex to be unstable. The polymerase keeps starting and releasing short RNAs (abortive initiation) until the transcript is long enough to stabilize the complex and allow sigma to detach.
During bacterial transcription, when does RNA polymerase release from the DNA template?
Once termination is reached
RNA polymerase stays bound to the DNA through initiation and elongation. It only releases the DNA when it encounters a termination signal (either Rho-dependent or Rho-independent) marking the end of transcription.
In bacterial transcription, which of the following features causes intrinsic (Rho-independent) termination?
a stem-loop (hairpin) structure followed by a stretch of uracils in the RNA
What happens in Intrinsic termination with G-C and A-U pairs ?
the RNA forms a G–C–rich hairpin that causes RNA polymerase to pause. The following U–A base pairs are weak, allowing the RNA transcript to separate from the DNA template and ending transcription.
In bacterial transcription, what is required for Rho-dependent termination to occur?
The Rho protein that binds the RNA
In Rho-dependent termination, the Rho protein binds to a specific site on the growing RNA and travels along it. When Rho catches up to RNA polymerase at a pause site, it causes the enzyme to detach from the DNA, ending transcription
A mutation occurs in the lacI gene that prevents the repressor from binding lactose. What will happen to lac operon expression in the presence and absence of lactose?
The operon is always off.
If the lac repressor cannot bind lactose, that means it will always remain bound to the operator (since lactose normally inactivates the repressor by binding to it and causing it to release). In the absence of lactose: The repressor is bound to the operator → transcription is off (normal state). In the presence of lactose: The repressor still cannot bind lactose, so it stays bound to the operator → transcription remains off.
E. coli is grown in a medium containing both glucose and lactose. What is the status of lac operon transcription?
Low
In the presence of glucose, ______ levels are low, so ____ does not activate the lac promoter effectively. Even though lactose inactivates the repressor, transcription occurs at a low level because the ____ activation is absent.
cAMP;
CAP;
CAP-cAMP