Molecular Bio Recitation Final Exam
1/38
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
39 Terms
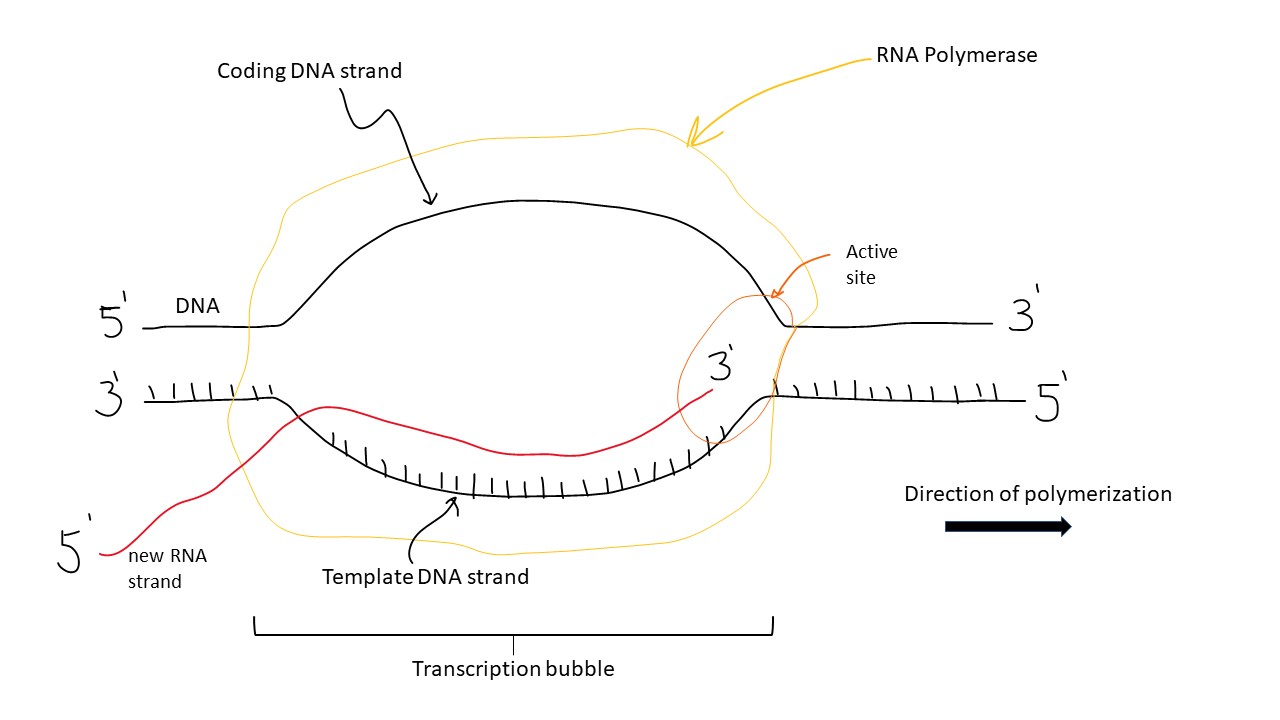
Transcription
the process of converting information from DNA into RNA
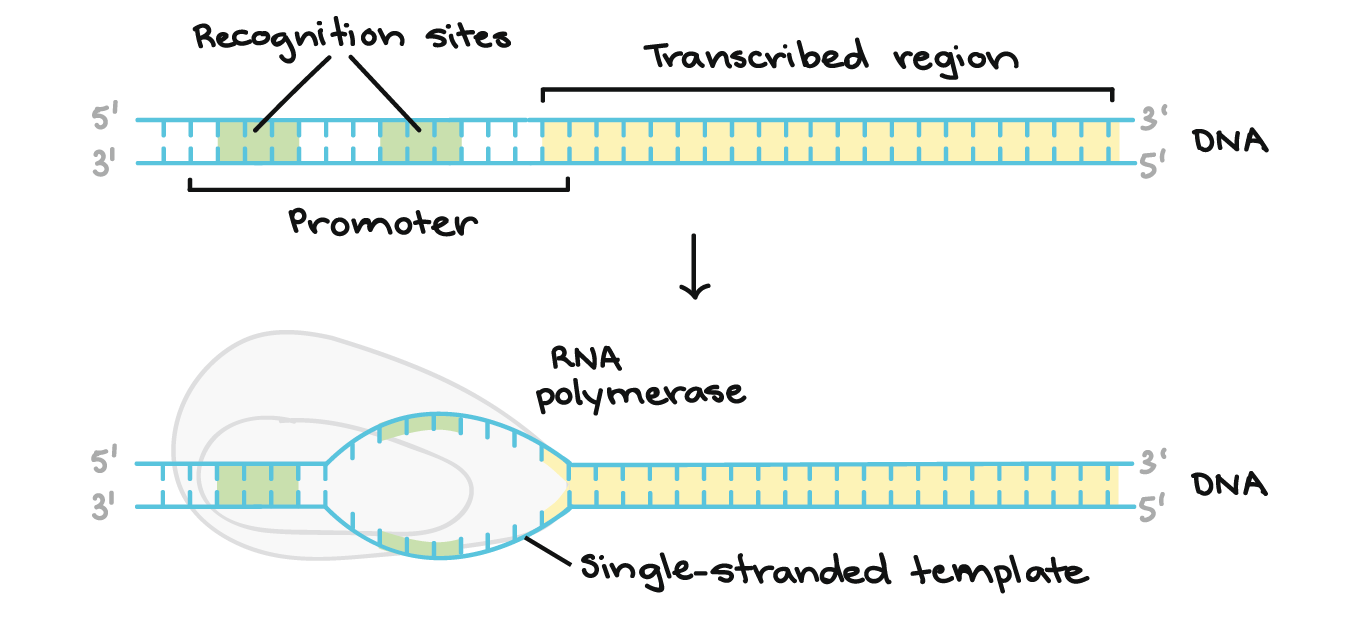
Initiation
the initial phase of gene expression where RNA polymerase binds to a specific DNA sequence, the promoter, and begins synthesizing an RNA copy of the gene
The most regulated step and rate-limiting step
• RNAP binds to promoter DNA
and unwinds ~14 base pairs surrounding the
transcription start site to create a RNAP-
promoter open complex (RPo)
• When the complex starts its initial synthesis
and producing short RNA strands that are
abortive it is called RNAP-promoter initial
transcribing complex (RPitc)
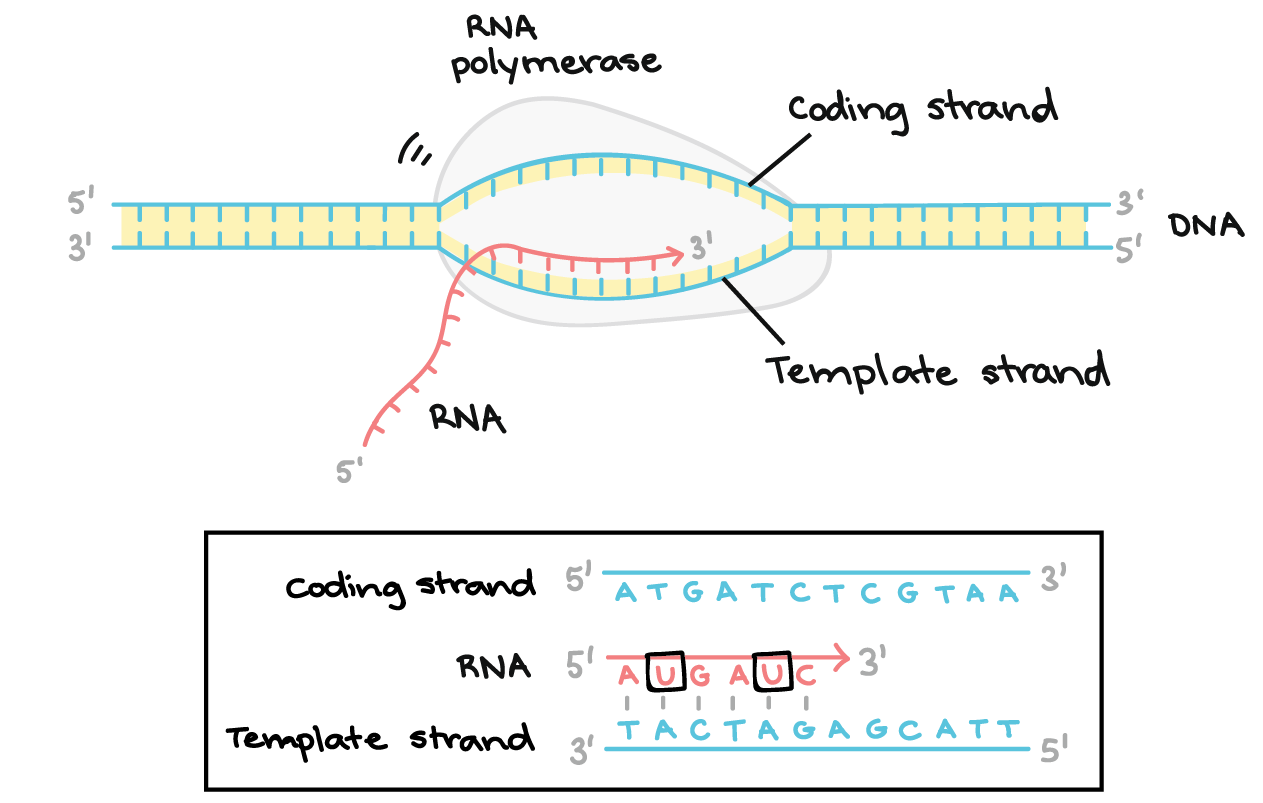
Elongation
the stage where RNA polymerase extends the nascent RNA transcript by adding new nucleotides in a 5' to 3' direction, following the template DNA strand
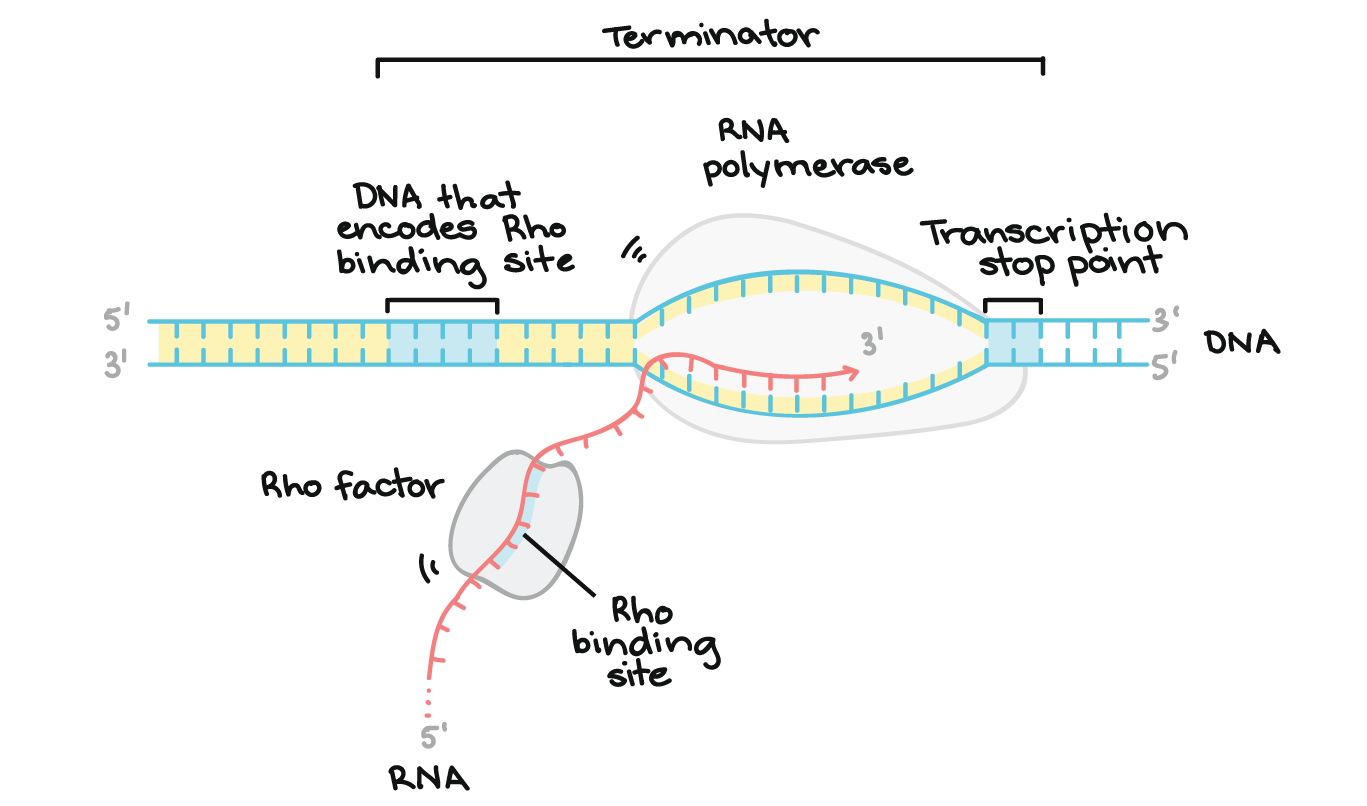
Termination
the process where RNA polymerase releases the newly synthesized RNA transcript and dissociates from the DNA template, marking the end of transcription
RNAP holoenzyme
The complete RNA polymerase enzyme, including core RNAP and the sigma (σ) factor, needed for promoter recognition and initiation.
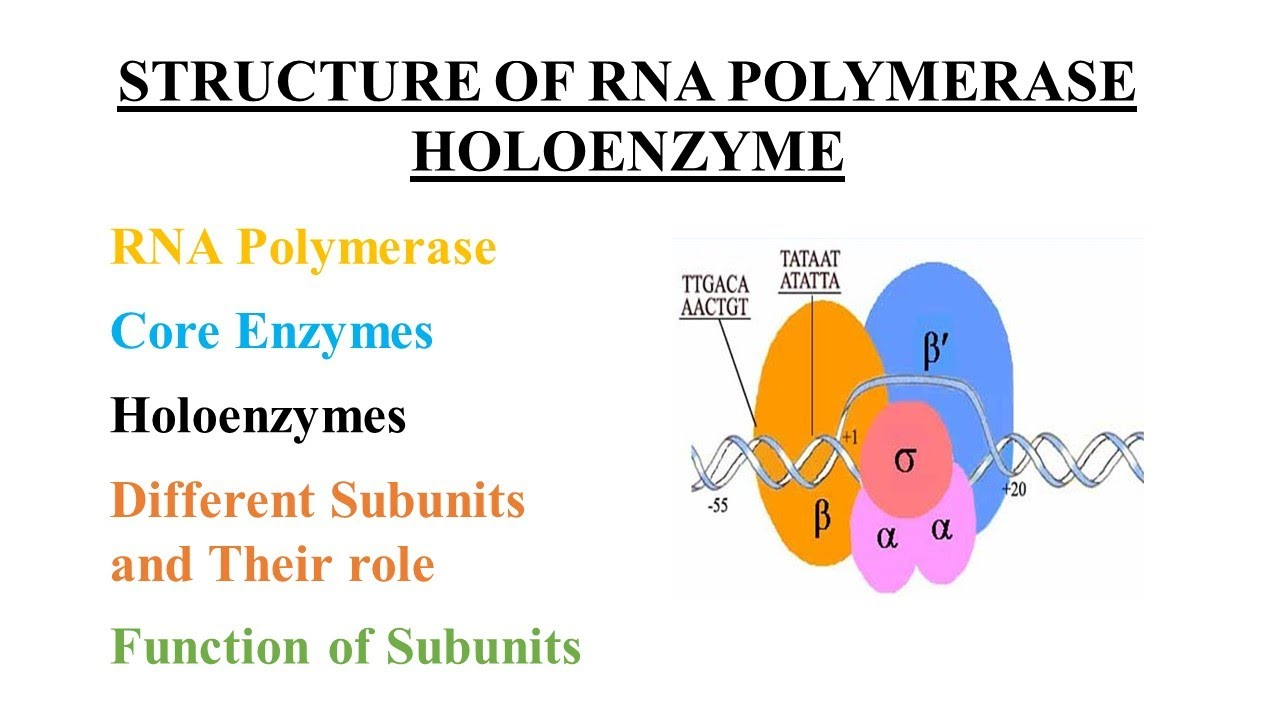
Sigma factor
a protein subunit of bacterial RNA polymerase that plays a crucial role in initiating transcription. It enables RNA polymerase to recognize and bind to specific DNA sequences called promoters, which mark the starting point of genes.
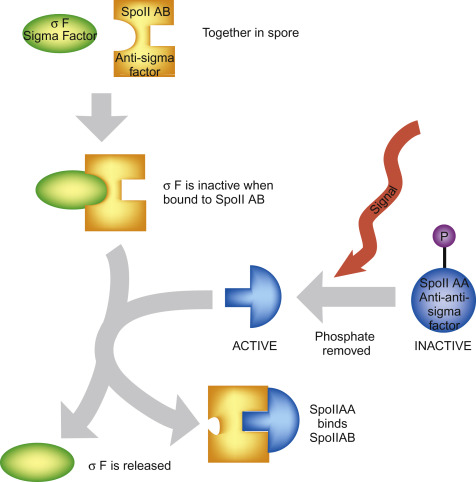
Translocation model
RNAP physically moves forward along the DNA with
each nucleotide it adds to the RNA chain.
•The DNA and RNA remain fixed inside the
polymerase, and the enzyme itself changes position.
•This resembles how RNAP moves during elongation.
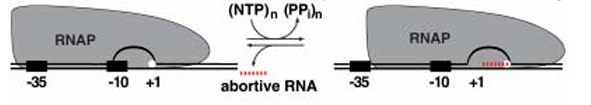
Inch-worming model
A portion of RNAP moves forward while the rest stays behind.
•After the forward part synthesizes some RNA, it pulls the rest of the polymerase along, like an inchworm
stretching and contracting.
•The active site shifts forward internally rather than through full translocation.
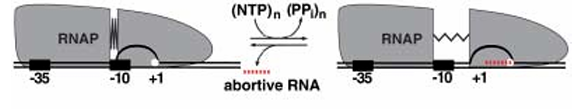
Scrunching model
RNAP stays fixed at the promoter region.
•Instead of the polymerase moving, it
pulls DNA into itself, causing the
downstream DNA to compress or
“scrunch.”
•This builds up tension (or stress) inside
the DNA-RNAP complex, which can later be released when RNAP breaks free to enter elongation.
FRET
Förster Resonance Energy Transfer
• Allows for real time observation of molecular movements at a
nanometer scale
• They fluorescently labeled DNA to track its changes in length and
position during transcription
• Applied single –molecule techniques to directly test models of transcription
Promoter escape
the final step of transcription initiation, where RNA polymerase detaches from the promoter region and transitions into the elongation phase. This involves releasing the polymerase from its initial complex and allowing it to transcribe the gene.
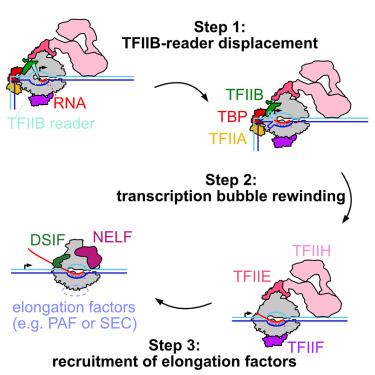
Abortive initiation
also known as abortive transcription, is a process where RNA polymerase binds to a DNA promoter and repeatedly synthesizes and releases short RNA transcripts before fully escaping the promoter
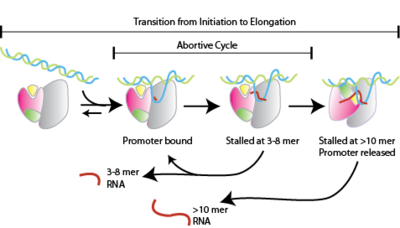
What are the three models proposed for RNAP active-center translocation during transcription initiation?
A: 1) Transient excursions – RNAP temporarily moves forward and backward.
2) Inchworming – The active center of RNAP moves independently, then retracts.
3) Scrunching – RNAP stays fixed and pulls in downstream DNA, creating tension.
What is abortive initiation during transcription?
A phase where RNAP repeatedly synthesizes and releases short RNA products (~2–9 nt) without transitioning to elongation.
What is a stressed intermediate in transcription initiation?
A transient state in which DNA scrunching creates unwinding and compaction stress, storing energy used for promoter escape or abortive release.
fluorophore
A fluorescent chemical used to label molecules and monitor interactions via fluorescence.
RPo
RNAP-promoter open complex
RPitc
RNAP-promoter initial transcribing complex
RDe
RNAP-DNA elongation complex
What is E*?
The observed efficiency of donor-acceptor energy transfer, indicating molecular proximity.
-Increased E* indicates closer proximity
shorter donor-acceptor distance → closer proximity..
What does R represent in this study?
The distance between donor and acceptor fluorophores, used to infer structural changes in RNAP-DNA complexes.
What is ALEX microscopy and its role in the experiment?
Alternating-laser excitation (ALEX) enables single-molecule FRET detection by rapidly alternating excitation wavelengths to measure energy transfer efficiency (E*) and stoichiometry (S).
Ribosome profiling
A method that uses deep sequencing of ribosome-protected mRNA fragments to monitor translation in vivo with subcodon (single-codon) resolution.
quantitative proteomics
measures protein abundance. Ribosome profiling complements it by revealing translation rates, which better predict protein levels than mRNA abundance alone.
Why is mRNA abundance not a perfect proxy for protein synthesis?
Because mRNA can be translationally regulated, leading to different protein output despite similar transcript levels.
Ribosome footprinting
A technique where ribosome-protected mRNA fragments (~30 nt) are isolated and sequenced to determine where translation is occurring.
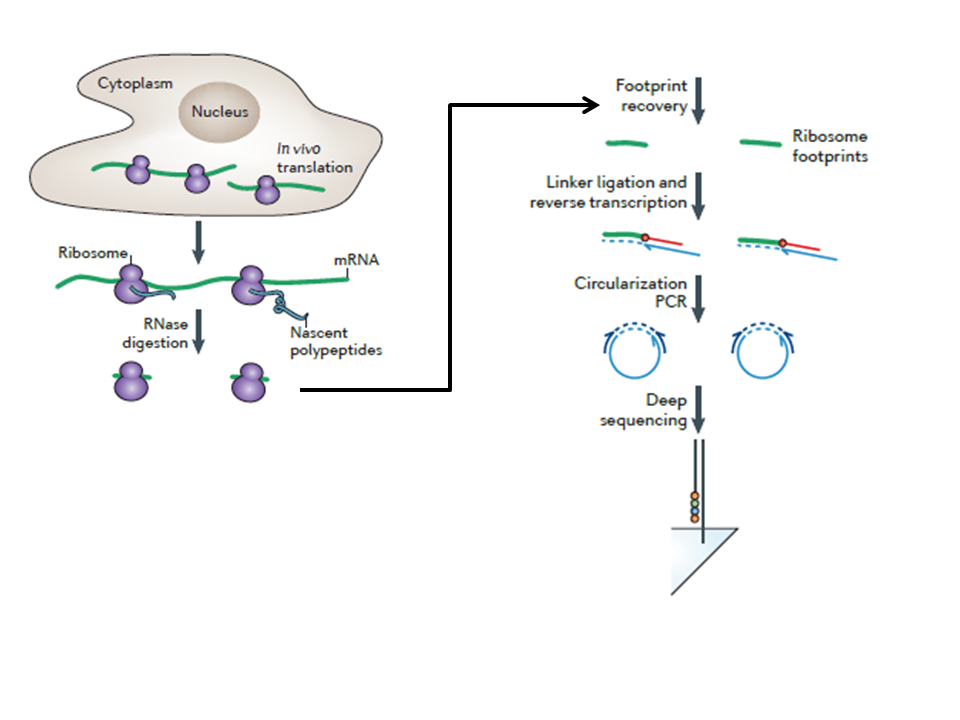
How does deep sequencing contribute to ribosome profiling?
It enables millions of short sequences to be read, allowing precise quantification and mapping of ribosome positions across the transcriptome.
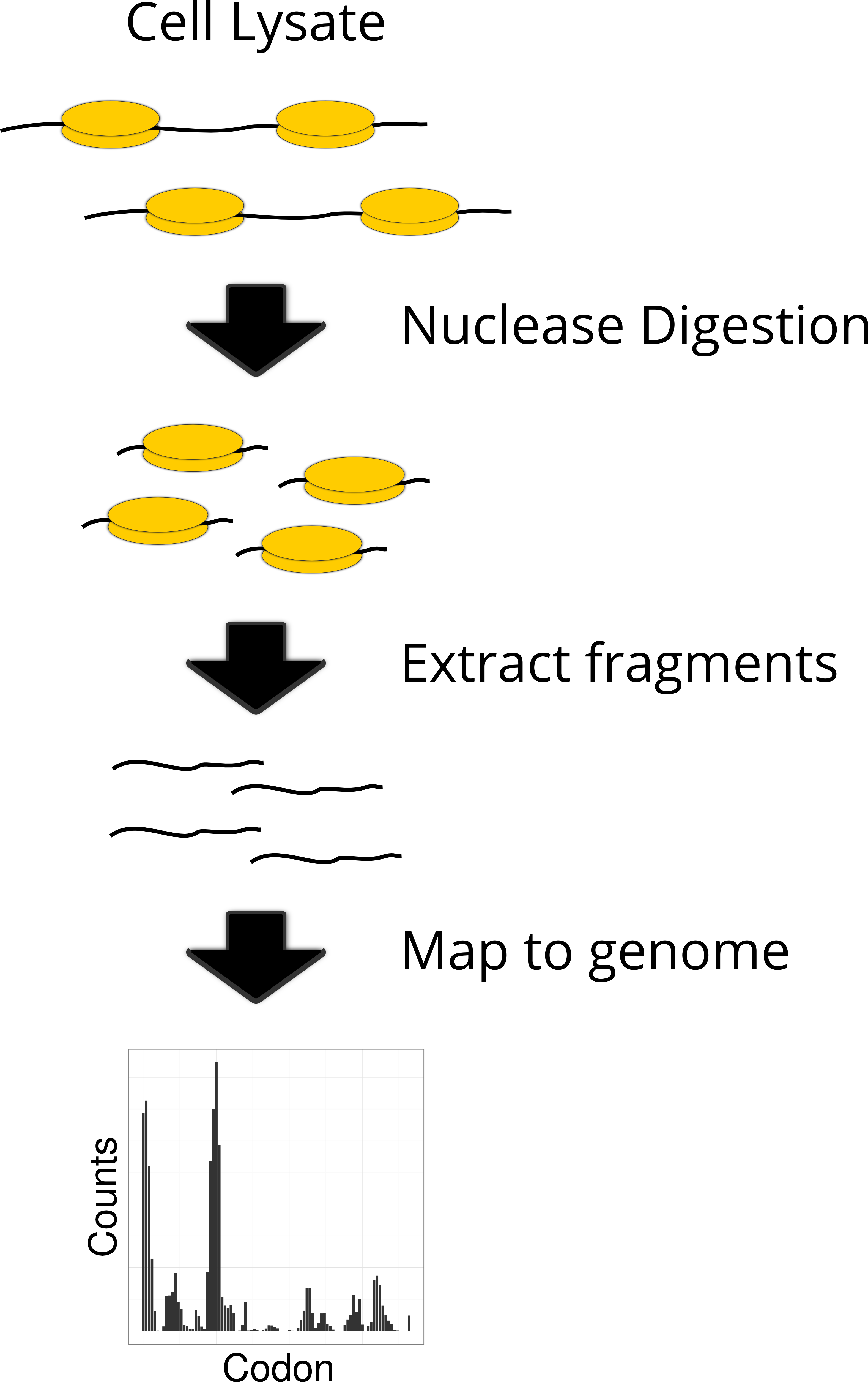
Deep sequencing library
A collection of DNA fragments derived from mRNA (or ribosome footprints) prepared for sequencing.
Polysome
An mRNA molecule bound by multiple ribosomes, indicating active translation.
Cycloheximide
A drug that freezes translating ribosomes on mRNAs, preserving ribosome positions at the time of harvest.
Ribosome density
A measure of how many ribosomes are bound per unit length of mRNA—used as a proxy for translation rate.
Donor fluorophore
absorbs light and becomes excited.
Acceptor fluorophore
if nearby to donor (typically within 1–10 nanometers), energy can transfer non-radiatively from the donor to the acceptor.
What structural region of the DNA was observed to "contract" during scrunching?
The transcription bubble (–15 to +15)
Subcodon resolution
particularly in ribosome profiling experiments, refers to the ability to identify and analyze the precise position of ribosomes on a mRNA molecule, down to the level of individual codons (three-nucleotide sequences) or even within those codons.
Which of the following observations supports subcodon resolution in ribosome profiling data?
Most 28-nt footprints start on the first nucleotide of a codon
-If most of those 28-nt fragments start at the first letter of a codon (not the second or third), that tells us:
Ribosomes are reading the mRNA in a very precise and consistent way
The technique is accurate enough to tell which exact codon is being translated
What is the significance of identifying footprints in 5′ UTRs?
Indicates active translation at upstream open reading frames (uORFs)
GCN4
gene in yeast (Saccharomyces cerevisiae) that encodes a transcription factor involved in the general control of amino acid biosynthesis.
-It activates many genes that help the cell synthesize amino acids, especially during nutrient stress (like starvation).
Under normal (nutrient-rich) conditions:
GCN4 mRNA is present, but its translation is repressed.
This repression is controlled by 4 upstream open reading frames (uORFs) in the 5′ UTR.
Ribosomes translate these uORFs and then fall off before reaching the GCN4 coding sequence.
Under starvation conditions:
A stress response alters translation (mainly via eIF2α phosphorylation).
Ribosomes skip the repressive uORFs and reinitiate at the GCN4 main coding sequence.
As a result, GCN4 protein is made, and it turns on genes needed for amino acid production.
What role do non-AUG start codons play in the study's findings?
They show that translation initiation is more flexible than previously thought