Prokaryotic Diversity Exam 2
1/265
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
266 Terms
what is the largest and most metabolically diverse phylum of Bacteria
Proteobacteria
explain the diversity of Proteobacteria
- contains species capable of phototrophy, chemoorganotrophy, and chemolithotrophy
- can be facultatively aerobic, microaerophilic, or stricly anaerobic
six classes of proteobacteria
alpha, beta, delta, gamma, epsilon, zeta
many species of proteobacteria have similar metabolisms (ex: methylotrophy or NH3/NO2- oxidation). what does this suggest?
suggests that HGT has been important in the evolution of proteobacterial metabolic diversity
for proteobacteria to grow autotrophically, what do they need BESIDES ATP?
NAD(P)H for CO2 fixation
Thus, when growing on poor electron donors such as H2S, NO2-, or Fe2+, energy must be used to drive electrons ______ a thermodynamic gradient
against

reverse electron transfer
energy-dependent movement of electrons against a thermodynamic gradient to form NADH
reverse electron transport steps
- electrons from AH2 are funneled into the respiratory chain and used to generate a PMF
- PMF used to generate ATP via ATP synthase
- PMF is partially consumed to reduce NAD+ to NADH
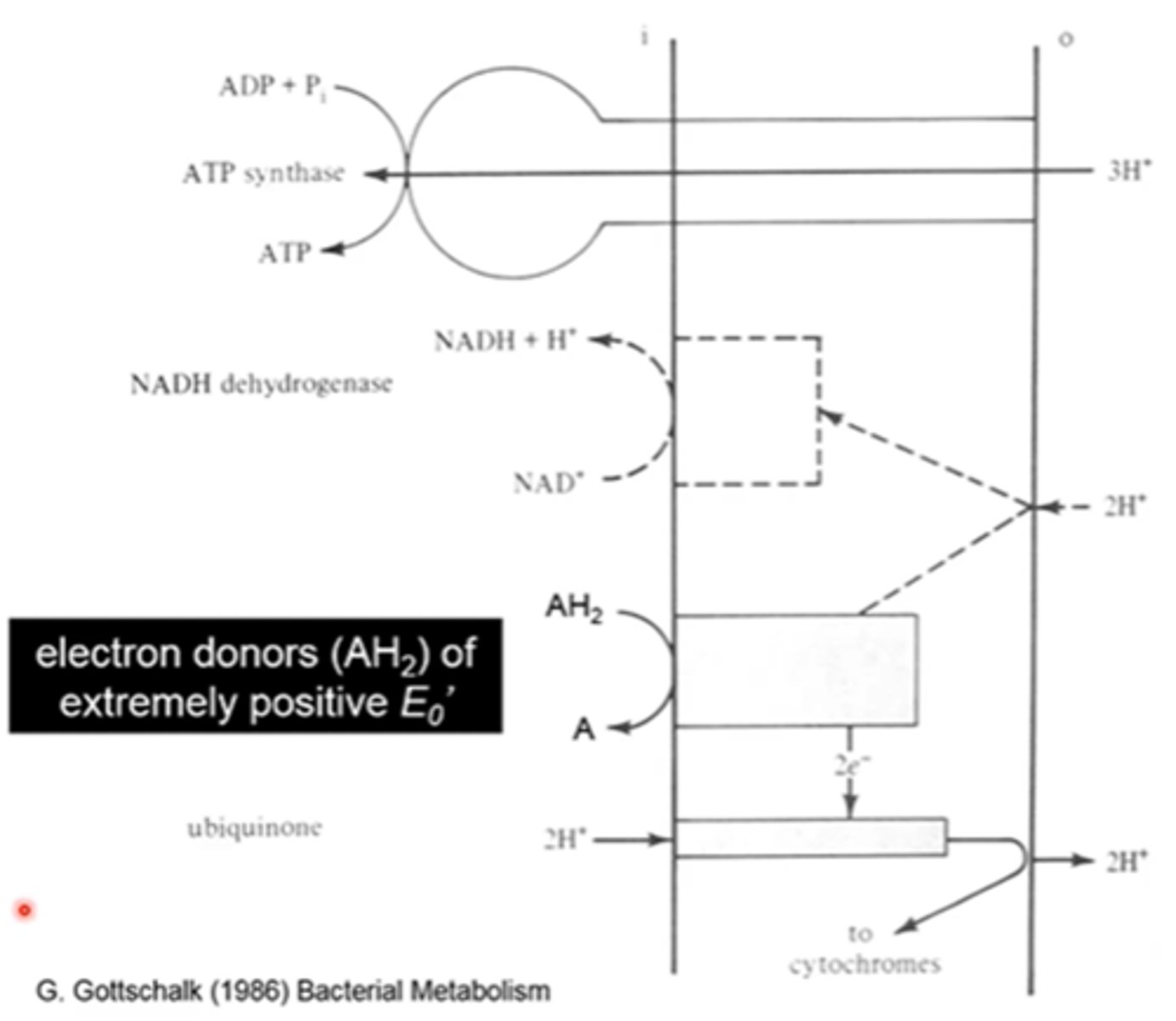
what do the dashed lines indicate in the picture in the last slide
reverse electron flow leading to NADH formation
NADH vs NADPH
- NADH is used in cell respiration
- NADPH is used in photosynthesis
reverse electron flow is used by ______ to generate _______ power
chemolithotrophs
reducing
what is reducing power required for
to fix CO2 for biomass
nitrosifying bacteria
- ammonia-oxidizing bacteria/archaea
series of step-wise reactions to:
1) oxidize ammonia into hydroxylamine
2) oxidize hydroxylamine into nitrite
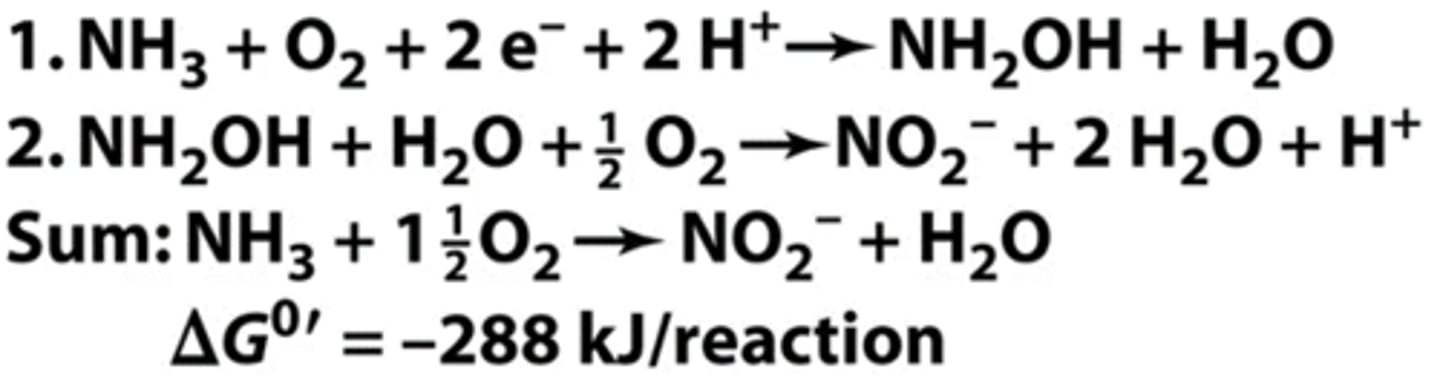
do nitrosifying bacteria require oxygen
yes (obligate aerobes)
nitrifying bacteria
- nitrite-oxidizing bacteria
oxidize nitrite to nitrate
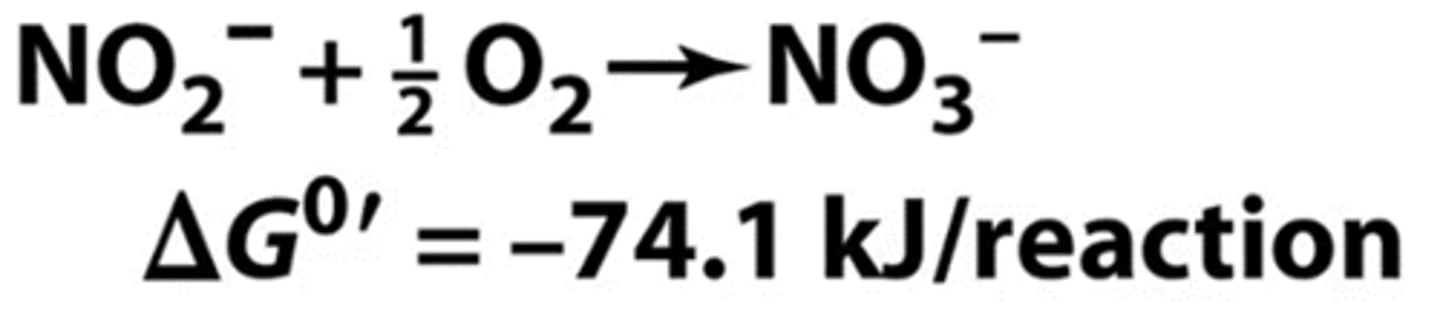
in nitrifying species, where are the enzymes of nitrification located
in the folded internal membranes
what are the enzymes of nitrification and their functions
- ammonia monooxygenase (NH3 --> NH2OH)
- nitrite oxidase (NO2- --> NO3-)
what is the purpose of the folded internal membranes of nitrifying species
to increase surface area for membrane reactions
what is the end product of nitrification
NO3 (nitrate)
nitrification process
soil bacteria oxidize NH4 to nitrite/NO2 then nitrate/NO3
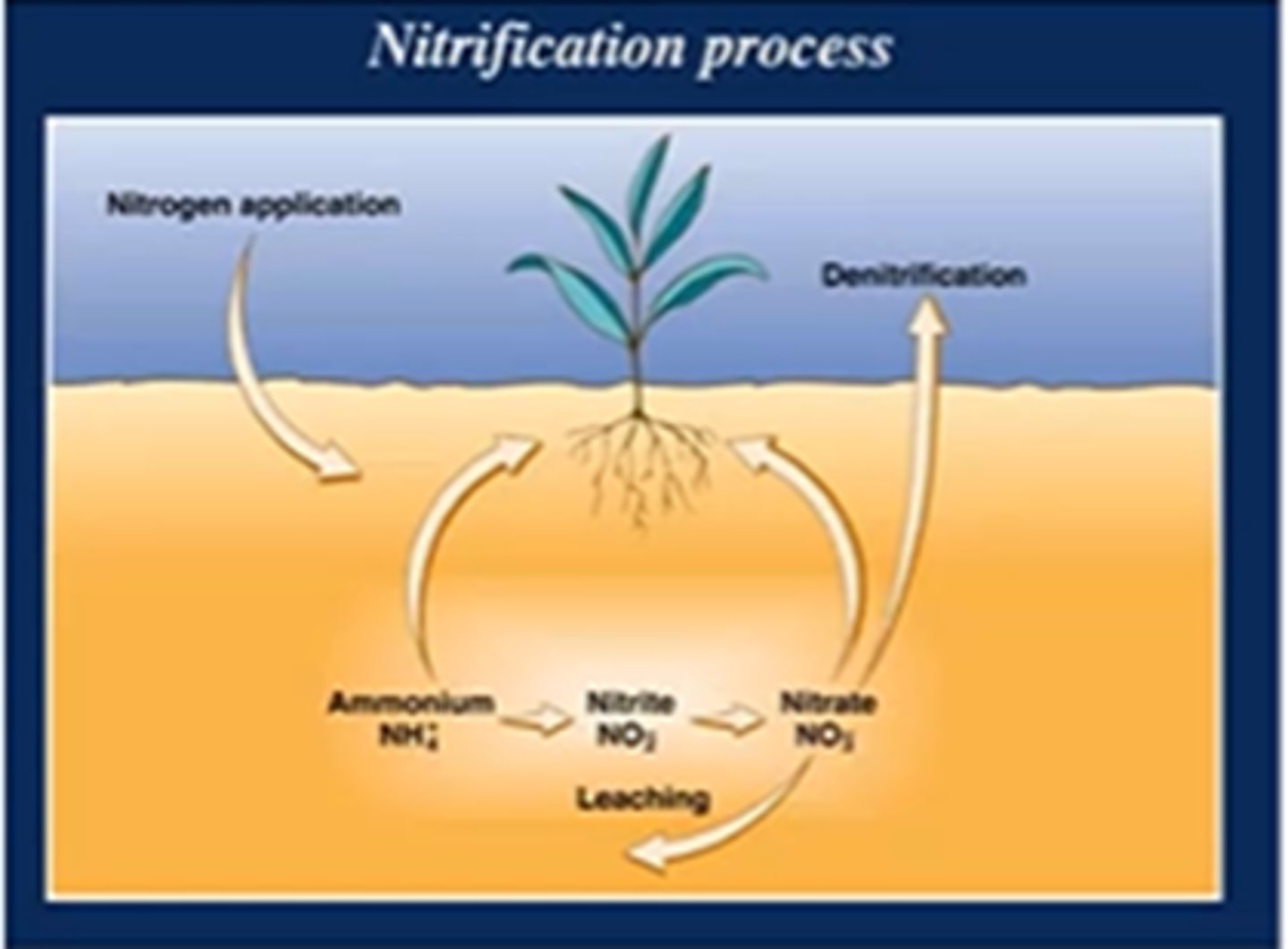
the end product of nitrification (nitrate) is the terminal electron acceptor for
de-nitrification
what is responsible for leaching of nitrogen fertilizers from agricultural ecosystems
activity of nitrosifying/nitrifying bacteria/archaea
populations conducting complete nitrification develop in soil and aquatic systems where ______ and ______ are present
ammonia
oxygen
where is nitrification typically found
oxic/anoxic transition zones with de-nitrifiers in close proximity
what were the first microbes that were shown to grow chemolithotrophically
nitrifying bacteria
what classes of the proteobacteria are nitrifying bacteria present in
4/6 classes (alpha, beta, gamma, delta)
two types of nitrifying bacteria
ammonia oxidizing
nitrite oxidizing
a genus beginning with "nitroso" means
ammonia oxidizers
what classes of proteobacteria are ammonia oxidizers found in
beta
gamma
a genus beginning with "nitro" means
nitrite-oxidizers
what classes of proteobacteria are nitrite oxidizers found in
alpha
delta
oxidation of ammonia by ammonia-oxidizing nitrifying bacteria:
- what are the 2 key enzymes
- ammonia monooxygenase (AMO)
- hydroxylamine oxidoreductase (HAO)
what does AMO do
what does NH2OH do
what does AMO require
what does it do with these things
what are the electrons supplied by
for every _____ electrons generated in the oxidation of NH2OH, only ____ reach the terminal oxidase
- oxidizes NH3 to NH2OH
- diffuses across the cell membrane (no net charge)
- requires 2 electrons and 2 H+
- reduces 1 atom of oxygen to H2O
- 4 electrons are supplied by HAO in the oxidation of NH2OH to NO2-
- 4, 2
what does HAO do
oxidizes NH2OH to NO2-
oxidation of nitrite to nitrate by nitrifying bacteria:
- what is the key enzyme
- what does this enzyme do
- the electron traverses a short electron transport chain consisting of ___ and ___ type cytochromes
- generation of the PMF occurs through the activity of
- since nitrite is an extremely weak reducing agent for NAD+, how many molecules of it have to be oxidized to generate 1 molecule of NAD+
- why do these microbes grow so slowly
- nitrite oxidoreductase (NOR)
- oxidizes nitrite to nitrate
- a, c
- cytochrome aa3
- 5
- because the reduction of NO2- to NO3- only generates a small amount of energy
who first conceived the notion of chemolithotrophy? what was he working on
Sergei Winogradsky
- working with Beggiatoa
Beggiatoa
- what class of proteobacteria are they
- what type of bacteria are they
- what environments are they common in
- where are they most abundant
- what did Winogradsky note while he was studying them
- what was his hypothesis
- gamma
- sulfur-oxidizing bacteria
- common in H2S-rich environments
- most abundant in the oxic-anoxic interface
- Beggiatoa starved for H2S lost their sulfur granules
- hypothesized that H2S was being oxidized to S0 as an energy source

what is the most reduced form of sulfur
what is the most oxidized form of sulfur
H2S
SO4 2-
Beggiatoa:
- in the presence of H2S, what happens
- in the absence of H2S, what happens
- S0 is deposited with the cell
- S0 globules are oxidized to SO4 2-
how many different catabolic pathways are used by sulfur-oxidizing bacteria
3 different pathways (1 generates ATP through phosphate level phosphorylation)
what are the different catabolic pathways of sulfur-oxidizing bacteria
1) H2S --> SO4 2- using the Sox system
2) H2S --> S) --> SO3 2- --> SO4 2-
3) same as 2 but with addition of substrate level phosphorylation
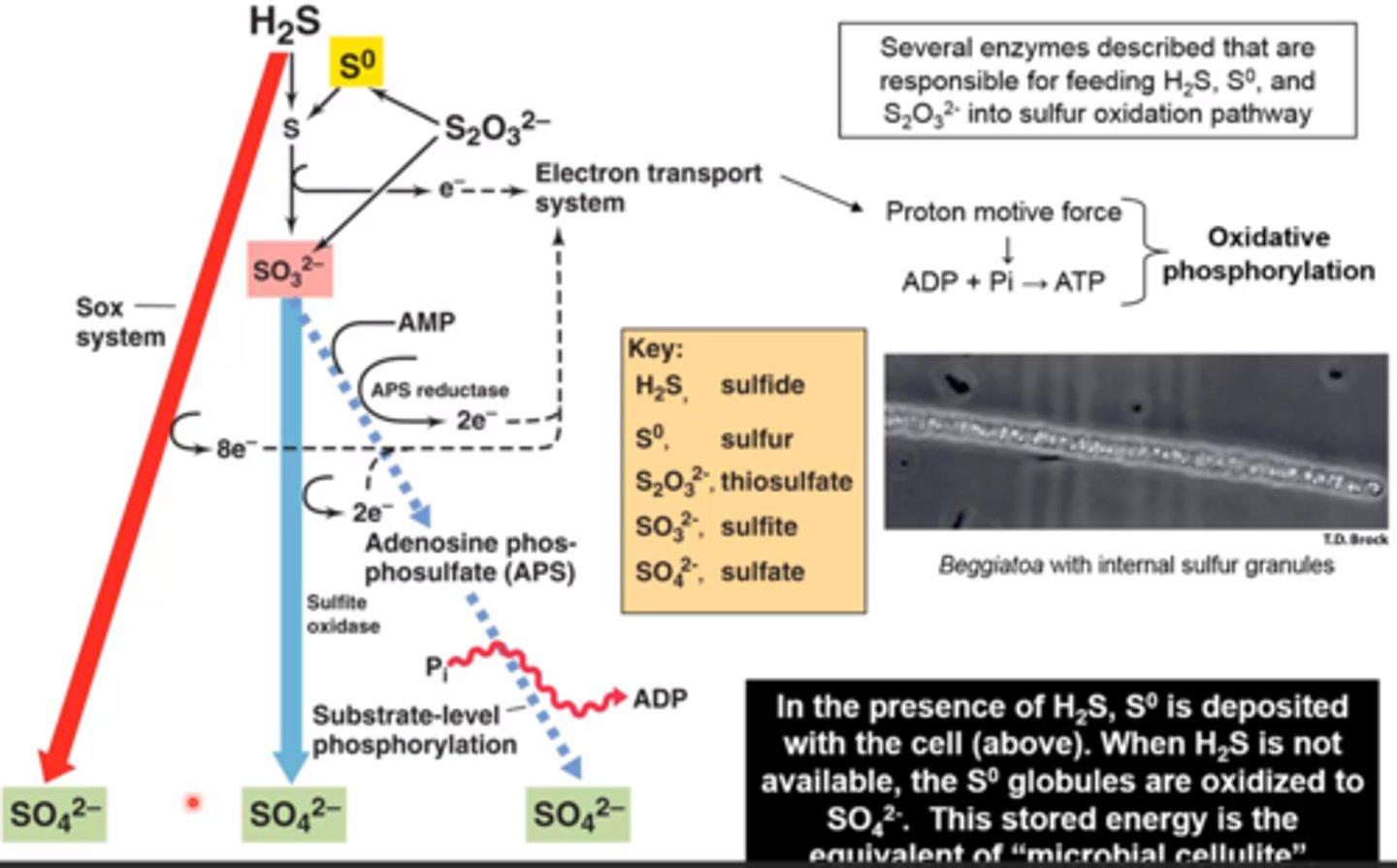
_____ is generated by energy-consuming reactions of reverse electron flow
why?
NADH
because the electron donors have a more electropositive E0' than NAD+/NADH
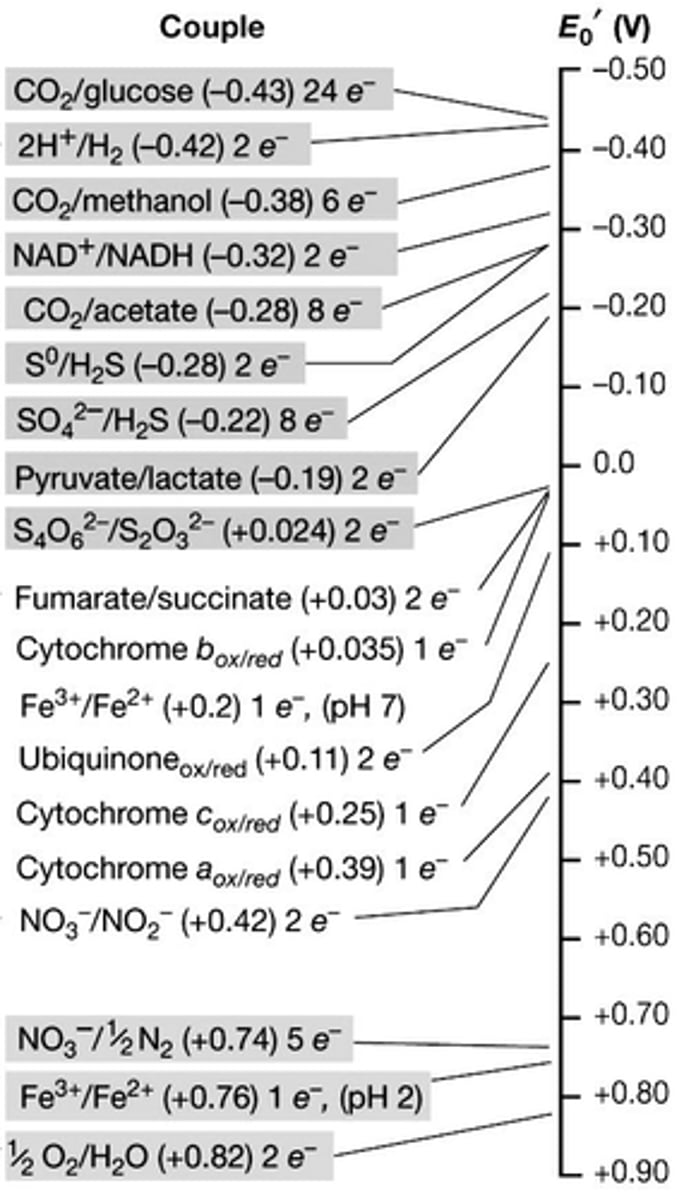
which of the following lithotrophic substrates does not require reverse e- flow to generate reducing power?
a) H2
b) H2S
c) NO2-
d) Fe2+
A
in most cases, the final product of sulfur oxidation is:
a) elemental sulfur
b) sulfate
c) thiosulfate
d) hydrogen sulfide
B
what metabolic advantage do cells have in storing the elemental sulfur biproduct from sulfur oxidation?
a) the byproduct can be used for other biosynthetic pathways that use sulfur, such as Rieske Fe-S proteins
b) the cells avoid producing transport energy waste to secrete the sulfur
c) sulfur decreases the intracellular acidification for non-acid-tolerant sulfide oxidizers
d) the byproduct serves as an electron reserve for subsequent oxidation
D
which of the following statements about the SAR11 and Pelagibacter ubique is incorrect?
a) they can assimilate sulfate (SO4 2-) as an S source to synthesize amino acids and thiol groups
b) none of the choices are correct
c) they possess the smallest bacterial genomes known
d) they are photosynthetic
B
the SAR11 were initially discovered by
a) using new methods of culturing marine microbes
b) the unique chlorophyll they possess
c) sequencing of their 16S rRNA genomes
d) morphological analysis of the cells in seawater
C
which of the following statements about this distribution of the SAR11 in marine ecosystems is correct?
a) they are the most abundant in deep waters during winter months
b) they are the most abundant in surface waters during winter months
c) they are the most abundant in deep waters during summer months
d) they are the most abundant in surface waters during summer months
D
DISCUSSION: regarding reducing power for autotrophic growth, what is the challenge faced by lithotrophs that use weak electron donors?
when an organism is using an energy source that is lower on the Redox tower (like Fe2+, NO2, or H2S), there's not much room for it to fall and generate energy
- these cells also require NAD(P)H to be able to fix CO2 which is higher up on the tower than the other substances
- because of this, these organisms have to use some of their energy to create NAD(P)+, which means that they grow and reproduce slower
DISCUSSION: what is an example of a lithotrophic substrate that does not require reverse e-flow to generate reducing power?
- in order to not undergo reverse e- flow to generate reducing power, a substrate listed ABOVE (more electronegative) NADH needs to be used (this includes glucose, H2, or methanol)
- since we're looking at a lithotrophic substrate, it will need to be an inorganic source (so it would need to be H2)
DISCUSSION: what microbial processes are exclusively responsible for the major oxidative branch of the nitrogen cycle?
nitrosifying and nitrifying bacteria are responsible for the major oxidative branch of the nitrogen cycle
- nitrosification is the process of oxidizing ammonia to nitrite
- nitrifying is the process of oxidizing nitrite to nitrate
DISCUSSION: for the nitrosifying bacteria (ammonia oxidizing) bacteria and archaea, what are the e- donor and acceptor used?
what about for nitrifiers (nitrite oxidizers)?
ammonia oxidizing (nitroso): donor is NH3, acceptor is O2
nitrite oxidizing (nitro): donor is NO2, acceptor is O2
DISCUSSION: where do the reactions of ammonia oxidation (nitrosification)/nitrification occur in the cell?
is this form of metabolism respiration or fermentation?
- the reactions take place across membranes in a regular electron transport chain, and some bacteria produce "organelles" with several layers of lipid membranes to have more surface area for the reactions to take place
- this is respiration because it uses an external molecule as an electron acceptor
DISCUSSION: how did observations of sulfur granules within Beggiatoa filaments lead Winogradsky to the concept of chemolithotrophy?
what are the e- donors and acceptors that chemolithotrophic sulfur bacteria use in their metabolic pathways?
- Winogradsky noted that Beggiatoa had sulfur granules when they were in H2S-rich water
- if H2S was depleted, the sulfur granules were lost over time (hypothesized that Beggiatoa was utilizing the elemental sulfur it had stored as an energy source in the absence of H2S)
- donors: H2S, S0, thiosulfate
- acceptors: O2, NO3, SO4
a survey of 16S rRNA genes in surface ocean waters revealed the presence of a novel microbial group. what was it
SAR11 cluster
when the 16S rRNA gene sequences of SAR11 were compared to those present in the database, what were they most similar to
members of alpha-proteobacteria
what % of prokaryotic rRNA in the Sargasso Sea is SAR11?
what % of total rRNA in the Sargasso Sea?
what % of all microbial cells throughout the oceans?
15%
12.5%
25%
SAR11 can account for as much as 50% of all cells in the Sargasso Sea
DOC
dissolved organic carbon
what is the main challenge of designing artificial media to mimic natural seawater
the accurate reconstruction of the complex composition of dissolved organic carbon (DOC) and trace elements
to mimic natural seawater, most researchers have relied on media that contains high concentrations of
organic carbon
what is widely used for culturing heterotrophic bacteria and has 170x more DOC than natural seawater
marine broth
what 2 other elements does marine broth have that are present in a lot higher concentrations than natural seawater
nitrogen
phosphorus
since most of the DOC in seawater is derived from _____ matter, the difference in usable organic carbon probably exceeds ____ orders of magnitude
recalcitrant
3
what is the rationale behind increasing carbon, nitrogen, and phosphorus in marine broth
does this work?
why/why not?
to increase microbial growth
- it actually has the opposite effect on many marine bacteria
- SAR11 exists in very dilute pools of organic matter in the ocean, grows best at very low nutrient concentrations, growth is inhibited by high nutrient levels
why is it important to cultivate microorganisms in the lab?
- examine how species are affected by changes in the environment and nutrients
- obtain genome sequences
- discover novel natural products (ex: antibiotic production)
- study cell-cell interactions
- link microbial diversity to metabolic potential
- understanding physiology yields information on the ecological rule of "uncultivable" species and their impact on biogeochemical cycling and human/animal health
what member of SAR11 was isolated in 2002 and was the first cultured of the SAR11 species
Candidatus Pelagibacter ubique
what class of proteobacteria is C. P. ubique
alpha
culturability of Candidatus Pelagibacter ubique using high-throughput sequencing vs. traditional plating on nutrient-rich agar media
culturability was 1.4-120x higher using high-throughput sequencing
in the Candidatus Palgibacter ubique cultures that were positive for growth, the cell titer never exceeds approx a million cells/mL. why is this significant
because that's approximately the same concentration that these microbes are found in seawater
what is a property of SAR11 that makes them successful in nutrient-poor conditions? why?
their small size
- provides high surface:volume ratio (enhances nutrient uptake)
- diffusion of substances throughout the entire cell volume is more efficient due to small size
what species has the smallest genome of any free-living organism and encodes the smallest number of ORFs known to a free-living organism
Candidatus Pelagibacter ubique
Ca. P. ubique genome size
despite its small genome, what DOES it contain
- 1.3 megabases
- contains nearly all basic functions of alpha-proteobacterial cells
features of the Ca. P. ubique genome
1) genes that are absent:
- genes that would confer alternate metabolic lifestyles, motility, or other complexities of structure and function
- non-functional or redundant DNA
- pseudo-genes, phage genes, or recent gene duplications
2) genes that are present:
- genes for carotenoid synthesis
- genes for retinal synthesis
- genes for proteorhodopsin
- median spacer size for intergenic DNA regions is 3 bases
what type of light-dependent proton pump does Ca. P. ubique have
light-dependent retinylidene proton pump
bacteriorhodopsin:
- what is it
- where is it found
- what species is it found in
- what does it do
- Pigmented protein
- found in the plasma membrane
- halophilic bacteria/archaea
- pumps protons out of the cell in response to light (light-driven proton pumps)
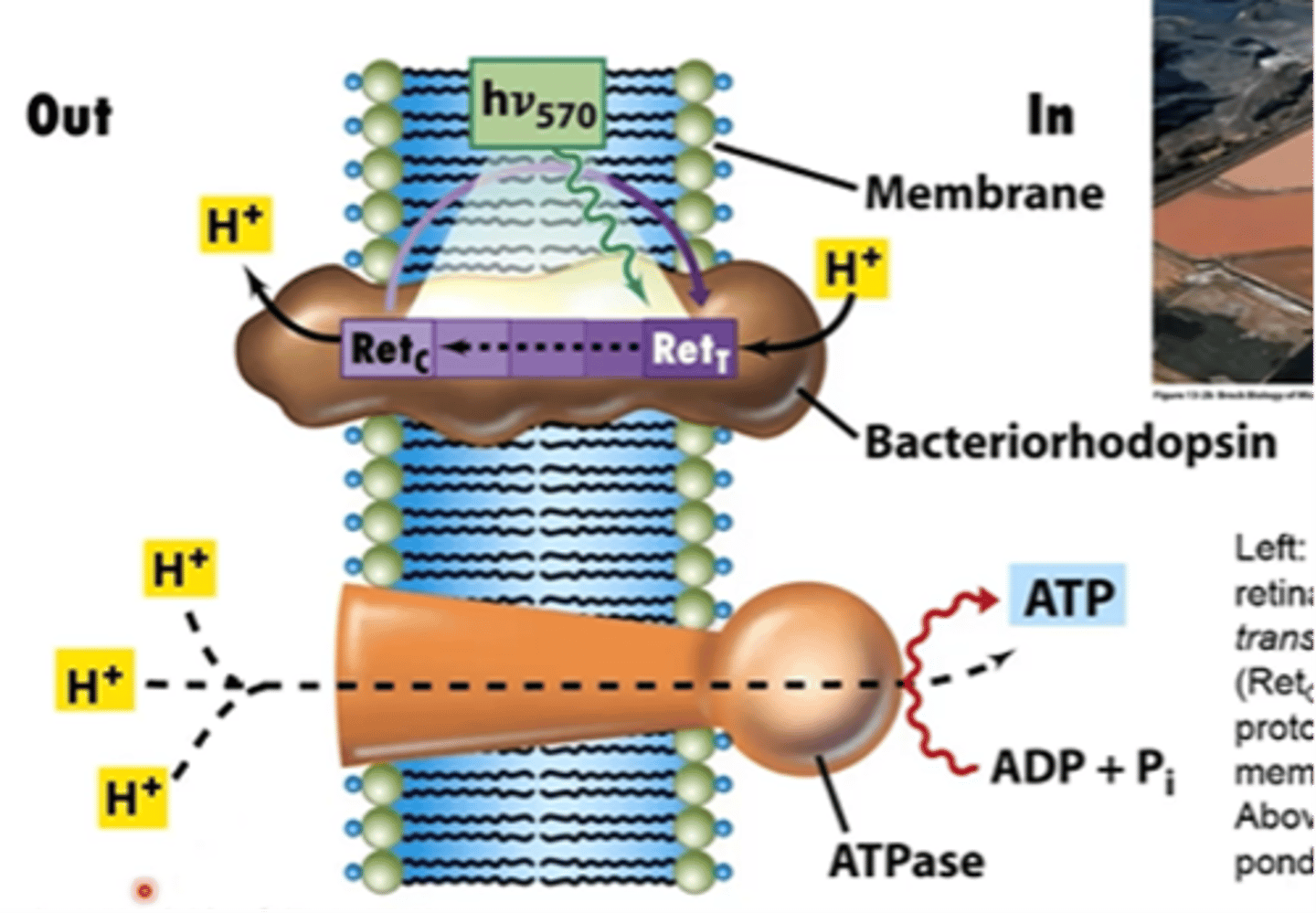
describe the Ca. P. ubique light dependent retinylidene proton pump
light converts the protonated retinal form of bacteriorhodopson from the trans form (Ret T) to the cis form (Ret c) along with translocation of a proton to the outer surface of the membrane
- this establishes a PMF
Proteorhodopsin:
- what is it
- what does it allow cells to do
- a form of rhodopsin
- allows cells to use light energy to drive ATP synthesis
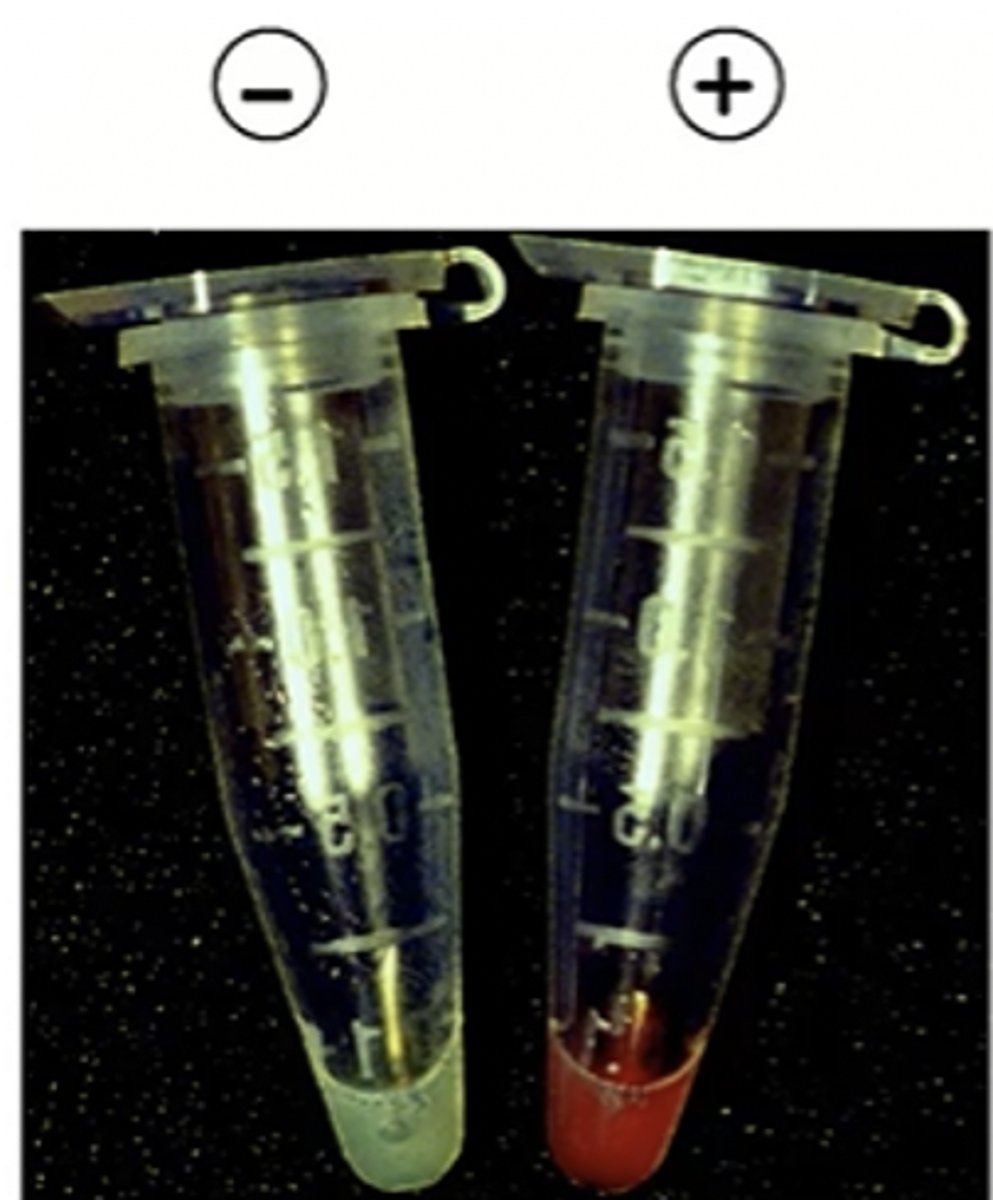
what color is an E. coli suspension that is expressing proteorhodopsin? what color are control cells? are these cis or trans forms of retinal?
expressing: red
control: green
both with all-trans retinal
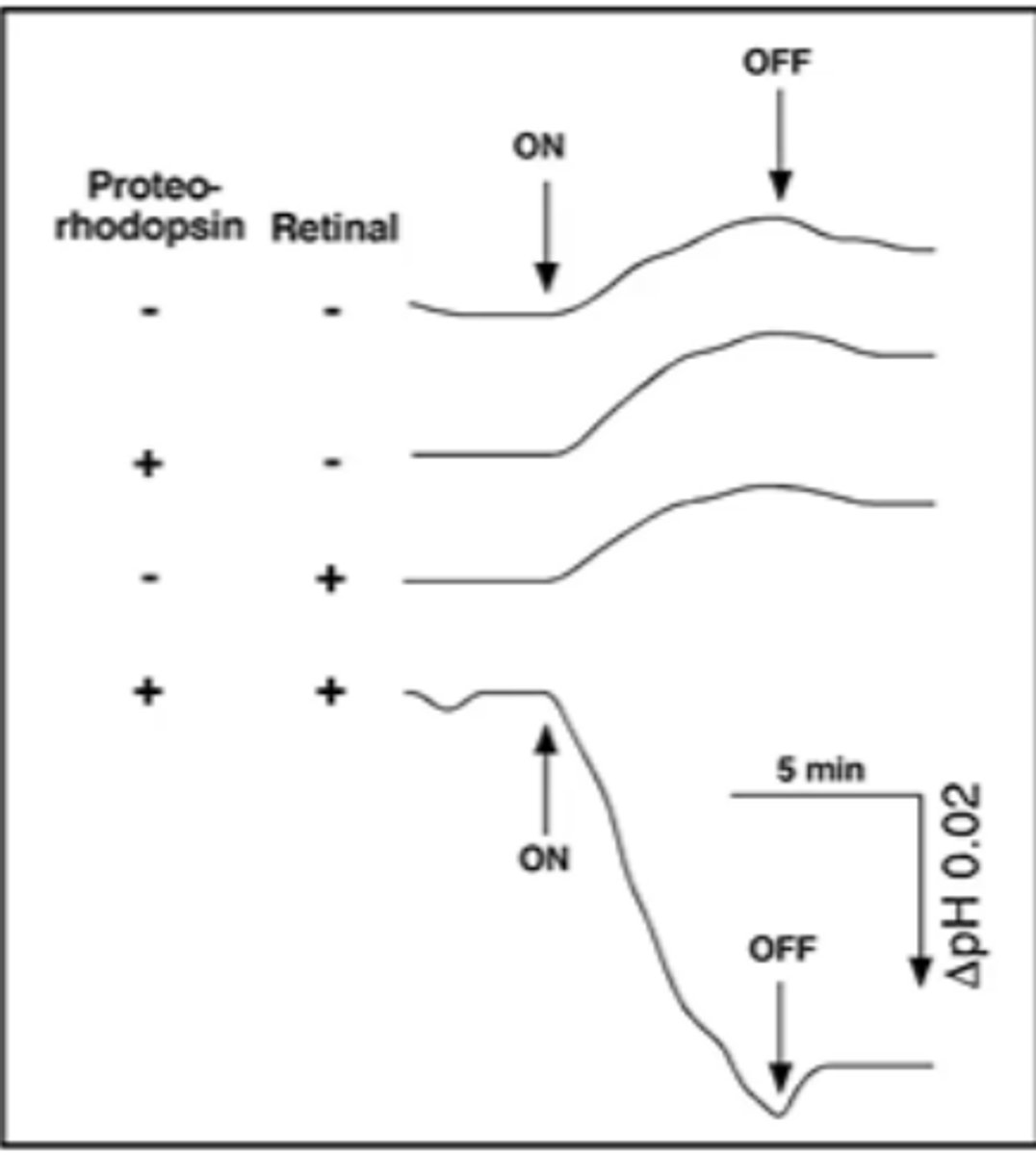
describe the light-driven transport of protons by a proteorhodopsin-expressing E. coli suspension in the following scenarios:
(Proteorhodopsin = P, Retinal = R)
1) P: -, R: -
2) P: +, R: -
3) P: -, R: +
4) P: +, R: +
beginning and cessation of illumination is indicated by the ON and OFF arrows
bacteria in the oceans generally derive their energy from
organic carbon
DISCUSSION: what are possible reasons why most microorganisms in nature have resisted culturing using conventional techniques?
- this is due to the highly diverse and dynamic ecosystem
- trying to produce an environment similar to ocean water is very challenging
- some species (like Ca. P. ubique) require lower nutrient levels
- high throughput sequencing can tell us more about metabolic potential and gene functionality (much easier now)
DISCUSSION: how was the SAR11 clade discovered? upon its discovery, why did many marine microbiologists begin studying this group intensely?
- discovered when a survey of 16S rRNA genes were collected and analyzed of surface level ocean water
- discovered it made up a large portion of the prokaryotic rRNA and that it was a distinct clade
- discovered that it was found all over the world and has many phylogenetic and physiological differences from microorganisms that had already been discovered
DISCUSSION: what parts of the ocean do the SAR11 tend to favor? what % of all microbial cells does SAR11 make up? what season do they dominate the ocean? what does this imply about their physiology and how it may be unusual?
- favor surface waters above 200m deep
- account for 25% of all microbial cells in surface waters
- dominate water particularly during the summer months
- their ability to thrive in low nutrient environments implies that they have unique physiology (small in size with increased surface:volume ratio and enhanced nutrient uptake, small but streamlined genome that assists in reducing the cost of replication and homeostasis)
DISCUSSION: when SAR11 was discovered, why was the presence of proteorhodopsin considered an unusual feature to be found in this microbe's genome? where is this protein located and what function does it serve?
how did SAR11 likely acquire proteorhodopsins?
- this type of protein was previously only found in Archaea
- found in the cell membrane and functions by driving a proton pump so bacteria can produce ATP from light in energy-poor conditions (useful when you live in a nutrient desert)
- likely acquired through HGT
DISCUSSION: what types of bacteria have smaller genomes than Pelagibacter ubique
only obligate parasites
DISCUSSION: "Marine broth 2216" was a standard media used by marine microbiologists for decades, yet it was not successful in allowing most microbes (and the most abundant ones) in the ocean to enumerate. when its chemical composition is compared to that of standard sea water, it is remarkably similar except for the elements C, N, and P. why do you think just these nutrients were increased to such high levels when this culture media was initially designed?
- these elements were increased to increase cell growth
- these elements are present in such low amounts in natural sea water that it's possible that the limited source of these elements could be a limiting resource in nature and is the reason that these organisms can't grow very fast
- it was thought that these were limited but required nutrients (if they were acquired then growth would increase)
DISCUSSION: why did increasing the concentration of C, N, and P in the media enhance the growth of some (but not all) microbes?
- microbes are extremely diverse in their nutrient requirements
- increasing the concentration of these elements might facilitate growth for some organisms, but microbes that have evolved under conditions of low nutrient-density do not thrive under nutrient-rich conditions (such as SAR11)
DISCUSSION: how were the members of the SAR11 clade eventually cultured? how did it compare to "conventional" microbial media?
- cultured using high-throughput cultivation
- this was 1.4-120x more effective than traditional plating on agar in terms of culturability
DISCUSSION: explain the process of high-throughput cultivation
1) seawater is collected and analyzed via 16S rRNA
2) flow cytometry is used to enumerate the number of cells in the sample and allow for precise dilution
3) microwell is inoculated with 1-3 cells
4) after a 2-4 week incubation period, the positive growth in the wells can be screened with flow cytometry to determine the number of wells that have growth
5) in the wells where growth occurred, samples can be taken for subculturing (16S rRNA sequencing)
DISCUSSION: oceanic surveys have revealed isolates such as Pelagibacter ubique represent just one member of a lineage in the SAR11 that is composed of numerous subclades. What could explain how this diversity has been generated? and what do we know about the occurrence of these clades that yields clues as to their ecological relevance?
- how these subclades grow (different nutrient and light availabilities, recombination, HGT, and mutations)
- as a result, various subclades have formed and evolved to occupy different niches in the ocean based on different light availabilities, temps, and nutrient concentrations
- ecological relevance: 30% of the bacteria found in the surface of the ocean during summer months
DISCUSSION: since Pelagibacter is a heterotroph, why might it care about the presence of light?
because it lives in a nutrient-poor environment, it is capable of utilizing light as an energy source when necessary using a light-driven proteorhodopsin proton pump when respiration is not available
where do methylotrophs grow
on compounds with no C-C bonds as a sole source of carbon and energy
methylotrophs vs methanotrophs
methylotrophs: use organic compounds or methylated compounds
methanotrophs: only use CH4 (methane)
most methylotrophs/methanotrophs are obligate _____
aerobes
two types of proteobacterial methylotrophs
type 1 (gamma proteobacteria)
type 2 (alpha proteobacteria)
type 1 methylotroph membranes
internal membranes arranged in bundles of disc-shaped vesicles
type 2 methylotroph membranes
paired membranes that run along the periphery of the cell
what is the purpose of the membranes in methylotrophs
same as in nitrifying bacteria (contain enzymes which the organisms use to conduct respiratory processes)
explain how methanotrophs generate a PMF
what is the terminal electron acceptor
what type of conditions does this process require
- oxidize CH4 to CH3OH via methane monooxygenase (MMO)
- use electron transport in the membrane to generate a PMF
oxygen
requires aerobic conditions
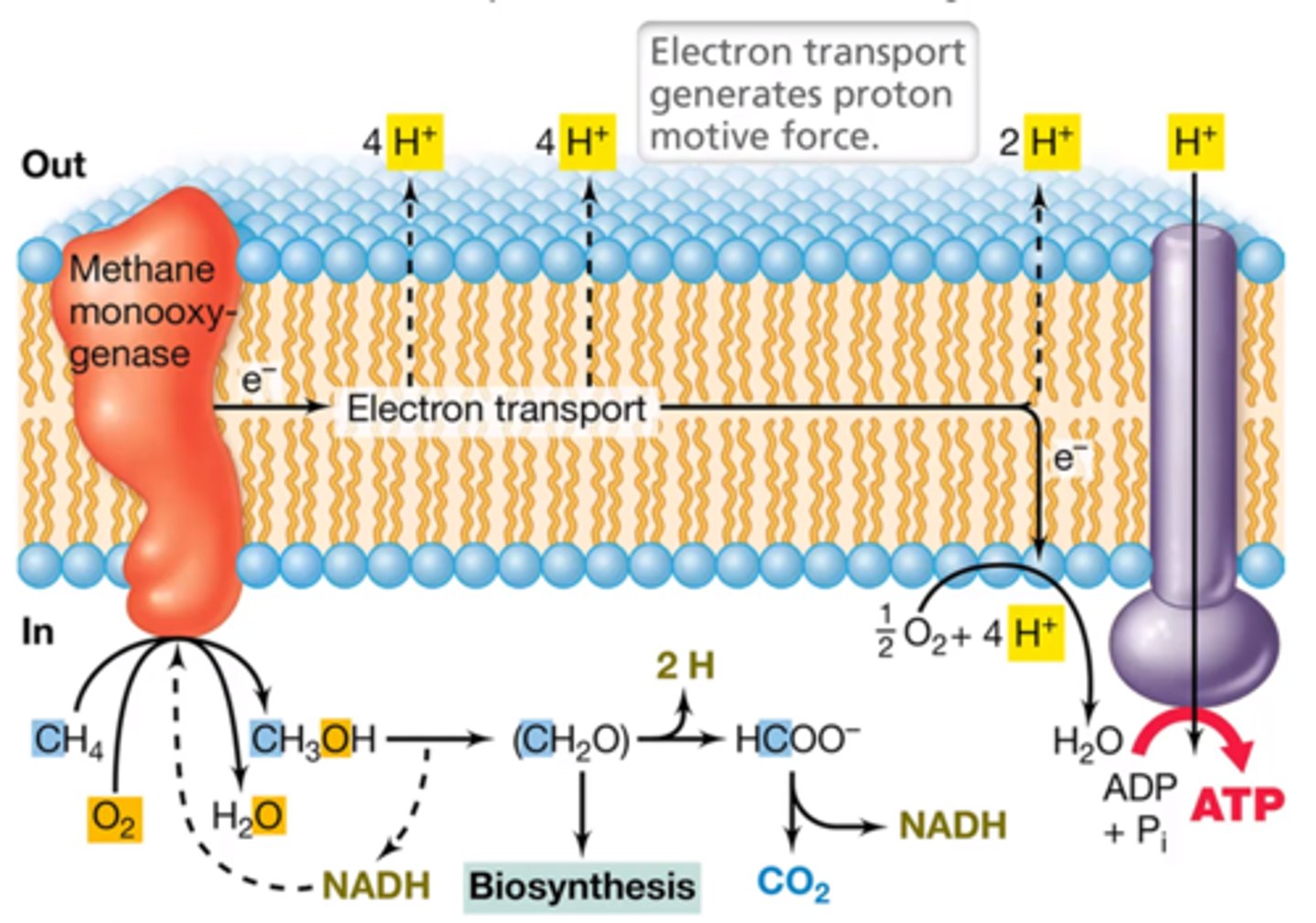
what are two types of MMO and what dictates which type is possessed?
soluble
membrane-associated
depends on the species