Biochem Lec 10-Enzymes II: Selectivity and Kinetics
1/32
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
33 Terms
What does Michaelis-Menten Kinetics define?
Defines in quantitative terms how enzymes behave and catalyze reactions under cell conditions, specifically how enzyme activity depends on substrate concentration.
The rate of formation and breakdown of the ES (enzyme-substrate) complex are governed by what?
k1, k-1, and kcat

What equation/relationship expresses the rate of ES formation?
Rate of ES formation= k1[S][E]
What equation/relationship expresses the rate of ES breakdown?
Rate of ES breakdown= k-1[ES]+kcat[ES]
k-1[ES] is rate of ES dissociation back to E and S
kcat[ES] is rate of ES conversion to E and P
![<p>Rate of ES breakdown= k<sub>-1</sub>[ES]+k<sub>cat</sub>[ES]</p><p> k<sub>-1</sub>[ES] is rate of ES dissociation back to E and S</p><p>k<sub>cat</sub>[ES] is rate of ES conversion to E and P</p>](https://knowt-user-attachments.s3.amazonaws.com/82953a22-5373-4e52-a924-30e0ee2dd1b7.png)
What is the Steady State Assumption?
At any given time in an enzyme reaction:
Rate of ES formation=Rate of ES breakdown
The [ES] remains constant
Necessary for M-M kinetics
From this, the M-M equation can be derived
What is KM?
The Michaelis constant is an important component of enzyme behavior and measures the affinity (strong/weak binding) of an enzyme for its substrate.
([E][S]/[ES])=(k-1+kcat)/k1=KM
![<p>The Michaelis constant is an important component of enzyme behavior and measures the affinity (strong/weak binding) of an enzyme for its substrate.</p><p>([E][S]/[ES])=(k<sub>-1</sub>+k<sub>cat</sub>)/k<sub>1</sub>=K<sub>M</sub></p>](https://knowt-user-attachments.s3.amazonaws.com/10f0ef7f-dd56-4163-b615-8c9638579dcc.png)
What is V0?
The rate of product formation.
V0=kcat[ES]
What is the relationship between [ES] and [E]t when all enzyme is bound to substrate?
[ES]=[E]t
[E]t=total enzyme=[E]+[ES]
What is Vmax and how is it determined mathematically?
Maximum rate of reaction when the enzyme is fully saturated with substrate.
Vmax=kcat[E]t
What is the Michaelis-Menten Equation and what do each of the variables represent?
1.Vo is the initial rate of the reaction (moles P produced/sec)
2.Vmax is the maximum possible rate when all E is occupied with S (i.e., when [ES] = [E]total )
3. [S] is the initial substrate concentration
4. KM is the Michaelis constant, it is the [S] that gives ½ Vmax
![<p>1.V<sub>o</sub> is the initial rate of the reaction (moles P produced/sec)</p><p>2.V<sub>max</sub> is the maximum possible rate when all E is occupied with S (i.e., when [ES] = [E]total )</p><p>3. [S] is the initial substrate concentration</p><p>4. K<sub>M</sub> is the Michaelis constant, it is the [S] that gives ½ Vmax</p>](https://knowt-user-attachments.s3.amazonaws.com/ba31d39b-7035-4d45-8c71-8f06b75b7466.png)
What is the shape the plot of Vo vs. [S] from the M-M equation?
Hyperbolic
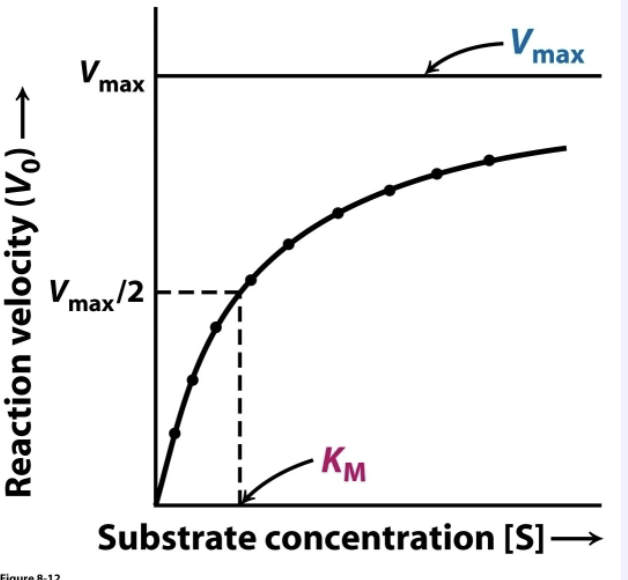
When does Vmax=Vo? Where is this represented on the plot?
Vmax=Vo when all enzyme molecules are bound with substrate. Represented by the curve’s asymptote.
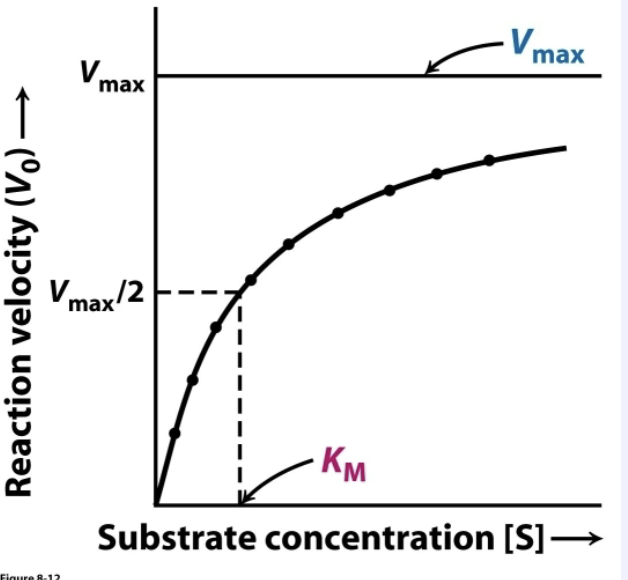
KM is the concentration of substrate ([S]) where _______
Vo= ½ Vmax
At very high [S] all enzyme molecules become saturated with S molecules. How does this impact the rate and reaction order? How does this relate to the equation for Vmax?
The rate no longer increases with more substrate
Zero order conditions
The equation for Vmax is Vmax = kcat [E]T → [S] is not part of the equation, so Vmax is not impacted by [S]
What is kcat and what does it represent?
“catalytic rate constant'“ or “turnover number”
It is the number of molecules of product P produced per sec per enzyme active site when all enzyme is saturated with substrate [S]
What is the inverse of kcat (1/kcat)?
The time needed for one enzyme active site to produce one product.
KM is a measure of the range of ____ where the enzyme is _____. It is often close to biological or cellular _____.
KM is a measure of the range of [S] where the enzyme is effective. It is often close to biological or cellular [S].
The ____ the KM the more effective the enzyme at low [S]
The lower the KM the more effective the enzyme at low [S]
How are Vmax and KM determined?
An enzyme kinetic experiment:
keep enzyme concentration constant
vary [S]
measure Vo (P produced/sec)
Plot data:
asymptote=Vmax
[S] that is ½ Vmax is KM
![<p>An enzyme kinetic experiment:</p><ul><li><p>keep enzyme concentration constant</p></li><li><p>vary [S]</p></li><li><p>measure V<sub>o</sub> (P produced/sec)</p></li></ul><p>Plot data:</p><ul><li><p>asymptote=V<sub>max</sub></p></li><li><p>[S] that is ½ V<sub>max</sub> is K<sub>M</sub></p></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/f6381d9c-bb1c-4a7a-8e22-8903e2bd3616.png)
What is an alternative way to obtain KM and Vmax?
Use the Lineweaver-Burk form of the M-M equation (“double reciprocal”)
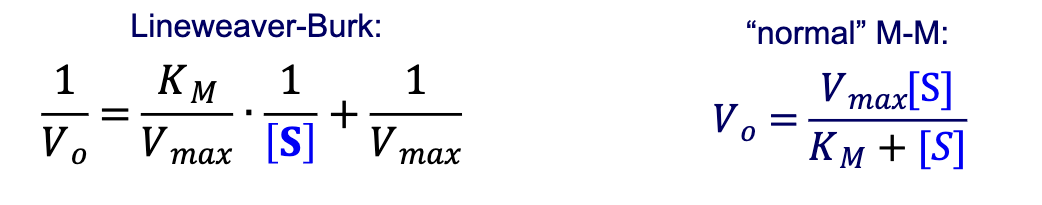
How does a plot using the Lineweaver-Burk form of the M-M equation differ? What is this plot called?
Lineweaver-Burk plot:
plot 1/Vo vs. 1/[S]
Describe a Lineweaver-Burk plot. What is its shape and why? What do the X/Y intercepts and the slope correspond to?
It is a plot of 1/Vo vs. 1/[S]. It has a linear shape because the Lineweaver-Burk equation—1/Vo=(KM/Vmax)*(1/[S])+(1/Vmax)—follows the y=mx+b format of linear functions.
X-intercept → -1/KM
Y-intercept → 1/Vmax
Slope → KM/Vmax
![<p>It is a plot of 1/V<sub>o</sub> vs. 1/[S]. It has a linear shape because the Lineweaver-Burk equation—1/V<sub>o</sub>=(K<sub>M</sub>/V<sub>max</sub>)*(1/[S])+(1/V<sub>max</sub>)—follows the y=mx+b format of linear functions.</p><p>X-intercept → -1/K<sub>M</sub></p><p>Y-intercept → 1/V<sub>max</sub></p><p>Slope → K<sub>M</sub>/V<sub>max</sub></p>](https://knowt-user-attachments.s3.amazonaws.com/58227e2e-5213-4592-83b6-45b5fe855050.png)
How do kcat and KM differ?
kcat → how quickly product is turned over once substrate is bound to the active site
KM → the effective substrate concentrations at which the enzyme works
What is the specificity constant? What two variables are used to determine it?
Describes the catalytic efficiency of an enzyme → how good the enzyme is at converting substrate into product
Describes the “catalytic efficiency” of an enzyme and kinetics when Vo < Vmax
Can be used to compare different substrates for enzymes
Determined by kcat/KM

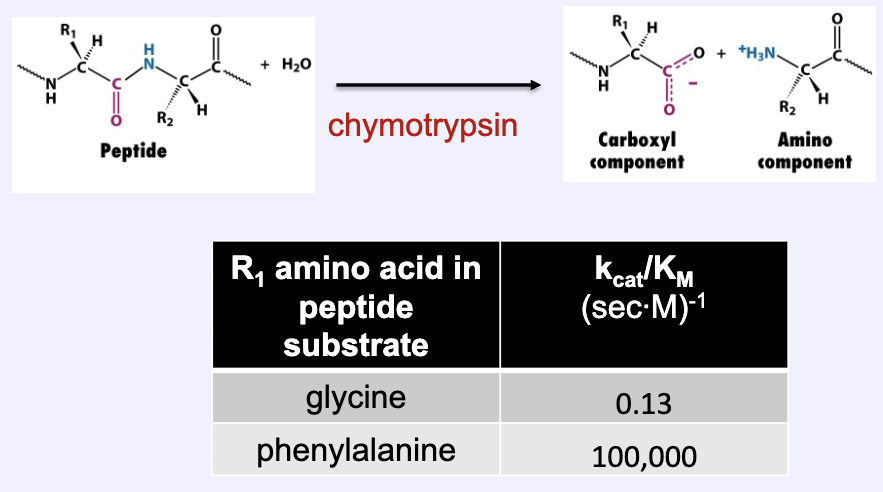
What is an example of how the specificity constant demonstrates enzyme efficiency/efficacy?
Chymotrypsin (a protease):
When R1 was glycine=0.13 (sec*M)-1
When R1 was phenylalanine= 100,000 (sec*M)-1
Chymotrypsin is more effective at peptide bond cleavage in the presence of a large/bulky/hydrophobic side chain than a small/flexible one → enzymes are specific to substrates
What is catalytic perfection?
Happens in extreme cases
Absolute efficiency in which every encounter with a substrate results in conversion to product (kcat/KM ≈ 109 sec-1M-1)
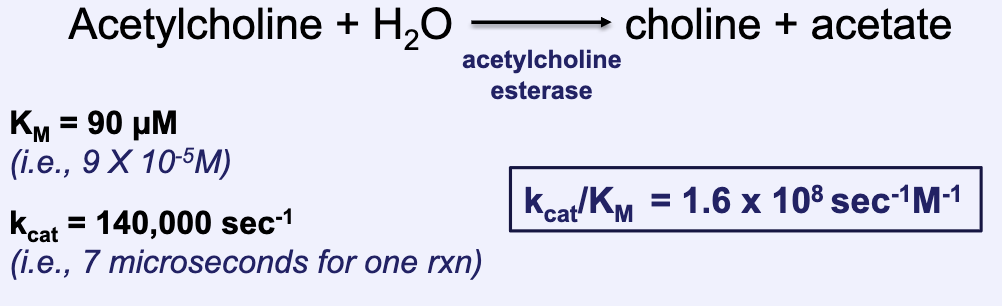
What limits the rate of reaction when catalytic perfection is reached?
The rate of reaction is limited by the time it takes for a S molecule to diffuse into an enzyme active site (i.e., diffusion limited)
M-M key concepts: What variables does the plot show a relationship between? What is its shape?
Hyperbolic relationship between rate of reaction (Vo) vs. [S]
M-M key concepts: describe Vmax and how it is determined mathematically.
Approached asymptotically
Rate when enzyme is saturated with substrate
Vmax=kcat[Et] → Et= E+ES
M-M key concepts:what is KM ?
M-M constant: KM = [S] at Vmax/2
M-M key concepts: what is kcat?
Turnover number → substrate molecules converted into product per second
kcat = Vmax / Et
M-M key concepts: what is the specificity constant and what limits it?
kcat/KM is catalytic efficiency. Better reflects rate under typical cellular conditions.
kcat/KM cannot be faster than diffusion-controlled encounter of enzyme and substrate (limit of ~109 s-1 M-1)
![<p>From the plot of velocity versus substrate concentration shown below ([E]T = 10<sup>-3</sup> μM), obtain the following parameters: a) V<sub>max</sub>, b) K<sub>M</sub>, c) turnover number (k<sub>cat</sub>), d) specificity constant (k<sub>cat</sub>/K<sub>M</sub>)</p>](https://knowt-user-attachments.s3.amazonaws.com/9094563c-050c-44a1-93ec-8984191463dc.png)
From the plot of velocity versus substrate concentration shown below ([E]T = 10-3 μM), obtain the following parameters: a) Vmax, b) KM, c) turnover number (kcat), d) specificity constant (kcat/KM)
a) asymptote → 6 μM min-1
b) point where [S]=1/2 Vmax → 0.5 mM
c) Vmax = kcat [E]T → kcat= Vmax/[E]T → kcat= 6 μM min-1/10-3 μM= 6×103 min-1= 100s-1
d) kcat/KM= 100 s-1/0.5 mM= 200 s-1 mM-1= 2×105 s-1M-1
![<p>a) asymptote → 6 μM min<sup>-1</sup></p><p>b) point where [S]=1/2 V<sub>max</sub> → 0.5 mM</p><p>c) V<sub>max</sub> = k<sub>cat</sub> [E]<sub>T</sub> → k<sub>cat</sub>= V<sub>max</sub>/[E]<sub>T</sub> → k<sub>cat</sub>= 6 μM min<sup>-1</sup>/10<sup>-3</sup> μM= 6×10<sup>3 </sup>min<sup>-1</sup>= 100s<sup>-1</sup></p><p>d) k<sub>cat</sub>/K<sub>M</sub>= 100 s<sup>-1</sup>/0.5 mM= 200 s<sup>-1</sup> mM<sup>-1</sup>= 2×10<sup>5</sup> s<sup>-1</sup>M<sup>-1</sup></p>](https://knowt-user-attachments.s3.amazonaws.com/abb53a7b-b3e9-4914-85fe-fe8531e22c9a.png)