BIOL 3000 Mutations
1/65
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
66 Terms
Genetic Material “Must haves”
Effective transfer between generations
Ability to store vast amounts of information
Information can be changed/mutable
Effective replication/high fidelity
Must be able to keep the mutation and replicate it
What does structure or configuration of a peptide chain confer?
The function of a protein
Where does the structure of a peptide chain come from?
From amino acids and how they interact with each other
If you change the sequence of the DNA or alter the structure of the polypeptide chain, then
You may alter the function of the resulting protein
Types of substitution point mutations
Transition mutant
Transversion mutant
Transition mutant
The exchange of a purine for a purine or a pyrimidine for a pyrimidine. Changing a singular base with a similar/like one.
Ex: ATG → GTG
Purines
Bigger
Adenine and Guanine
Pyrimidines
Smaller
Cytosine and Thymine
Transversion mutant
The exchange of nucleotides outside of a nucleotide family; the exchange of a purine for a pyrimidine.
Ex: ATG → T/CTG
Which happens more often: transitions or transversions? Why?
Transitions because the structure stays the same. Cells do not like using energy, so going from a base to a similar one takes less energy.
Types of insertions/deletions point mutations
Silent
Missense
Nonsense
Frameshift mutants
In-frame mutants
Silent mutations
The changing of one codon to a synonymous codon causing no change in the amino acid sequence of the protein. There is no change in gene expression.
“Degeneracy of Genetic Code”
Which nucleotide do silent mutations usually take place?
Generally, at the third nucleotide of a codon which causes no change in the amino acid coded for.
“A change in genotype causes no change in phenotype”
Missense mutation
The changing of one codon to a different codon, resulting in a change in the amino acid sequence of the protein. It does affect the amino acid.
Which nucleotide do missense mutations usually take place?
Generally, at the first nucleotide. If the first position is changed, then it will almost always change the amino acid.
How is the severity of missense mutations measured?
It typically depends on what amino acids are involved and where the missense mutation occurred
Nonsense mutations
The changing of one codon to a “STOP” codon, resulting in the premature stoppage of translation. It completely changes what the polypeptide is. The effect of the mutation depends on where it is. It makes the ribosome less sensitive to premature STOP codons.
Ex: Cystic fibrosis, Duchenne Muscular Dystrophy (DMD)
Frameshift mutation
The gain or loss of a nucleotide (or nucleotides) that result in the change in the reading frame of the codon. There can be a gain or loss of multiple. This can shift the reading frame, changing everything after the mutation.
It can result in a STOP codon, which would make it a nonsense mutation
Ex: Crohn’s Disease
In-frame mutation
The gain or loss of a nucleotide or trinucleotide set that does not change the reading frame of the codon.
This is a missense mutation.
Types of functional mutants of point mutations
Gain/loss of function mutants
Lethal mutants
Loss-of-Function Mutation
Results in a gene product with little or no functionality, amorphic. Most of the time, these phenotypes are recessive. There is a protein made, but the function is lost.
Gain-of-Function Mutation
Results in a gene product that has gained a new and abnormal function, neomorphic. These mutations are typically dominant.
Amorphic
The complete loss of a gene function
Neomorphic
A new or different function from normal
Lethal mutation
Mutation that leads to the death of the organism carrying the mutation. This is pretty bad, and can be any kind of mutation
Mutation in somatic cells
It can range from mild to severe, but is not passed to the next generation. Most of the time, induced mutations. They may pass down the “tendency” to have something.
Mutation in germ cells
Typically more severe manifestation because they can be passed along to offspring. It is something that can stay in the population.
Ex: breast cancer, hemophilia
Spontaneous mechanism of mutations
Replication errors
Chemical changes
Induced mechanisms of mutations
Environmental factors
Chemical interactions
Spontaneous mutations
Any mutation where no artificial factor or external regulator causes the mutation. Typically happens in replication errors.
Replication errors
Every time a cell divides; it must make an exact copy of 3 billion nucleotides to pass to daughter cells. 6 billion base pairs per diploid cell. The mutation rate of 1 per 100,000 bases.
120,000 mistakes EVERY time a cell divides.
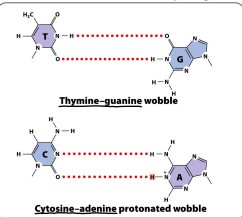
Results in the Non-Watson-and-Crick base pairing
Non-Watson-and-Crick base pairing
AKA “wobble”
Non-complimentary bases can pair due to the flexibility of DNA double helix which can accommodate slightly misshaped pairings.
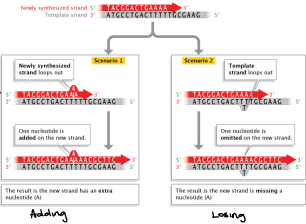
Slipped Strand Mis-pairing
Involves the denaturation and displacement of DNA strands that results in mispairing of complimentary bases.
Chemical changes
Mutations caused by normal chemical reactions that occur in the cell
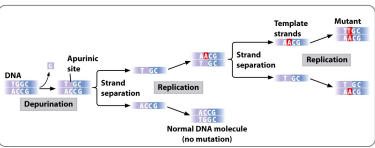
Depurination
A chemical reaction in which a β-N-glycosidic bond is cleaved by hydrolysis causing the release of an Adenine or Guanine from a DNA strand. Losing a purine.
Still having a backbone, just no base.
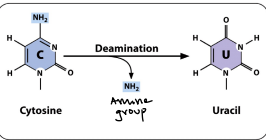
Deamination
The hydrolytic removal of an amine group from a nucleotide releasing ammonia and converting the nucleotide to another nucleotide. Losing an amine group. This is the most common single nucleotide mutation in DNA.
Types of induced mechanisms of mutations
Chemical interactions
Environmental factors
Chemical interactions
5-Bromouracil
5-BrdU
5-Bromouracil
A base analog or antimetabolite of uracil that can replace thymine in a strand of DNA
5-BrdU
The deoxyribose form
Used to study cancer cell proliferation as it is neither radioactive nor toxic to the cell
Environmental factors
Free radicals
UV light
Free radicals
Very unstable and quick reacting molecule that “steals” electrons from nearby stable molecules. They can be missing any number of electrons
UV Light
Causes pyrimidine dimers by the formation of covalent linkages localized on cysteine double bonds
Chromosome duplication
The duplication of a region of DNA that contains a gene
Causes of chromosome duplication
Ectopic recombination
Chromosome deletions
Small: less likely to be deletions
Medium: responsible for a number of genetic diseases
Large: often fatal
Transposable elements (TEs)
AKA “jumping genes” or transposons, mobile sequences of DNA that move (or jump) from one location in the genome to another that often generate some type of mutation when they move from one location to another on a chromosome and in evolutionary terms
It can increase the size of the genome
Might carry out some biological function, most likely regulatory one.
Thought to be junk DNA but now believed to make up about 40% of human genome
What was used to observe transposons?
Corn
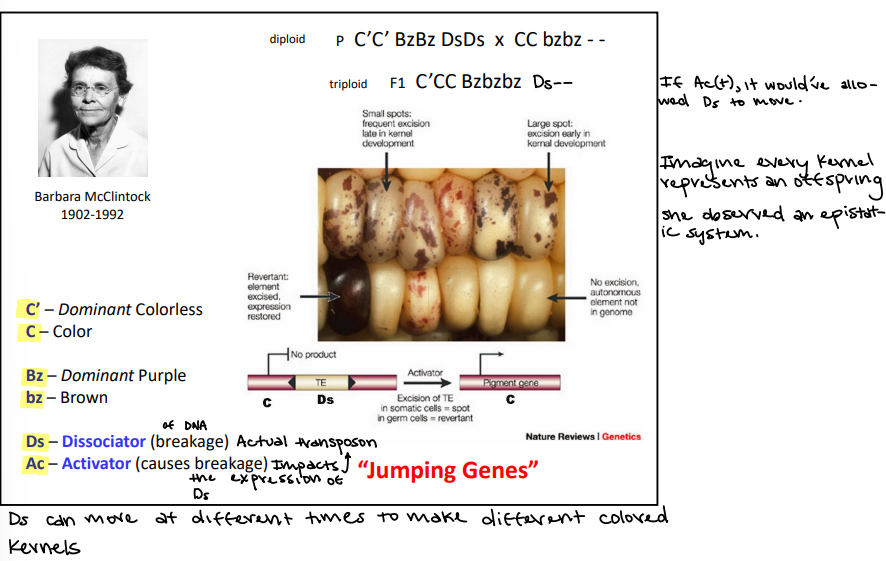
Ds
Dissociator (breakage of DNA), this is the actual transposon
It can move multiple different times
Ac
Activator (causes the breakage of DNA) and impacts the expression of the Ds
Class 1 TEs
Requires a reverse transcriptase (the transcription of RNA into DNA) in order to transpose
Retrotransposons
REtrotransposons = REquire reverse transcriptase
Class 2 TEs
Do not require reverse transcriptase in order to transpose
DNA transposons
Mechanism of Class 2 TEs
Autonomous Class 2 TEs encode transposase, DNA transposons
Moves throughout the genome in a “cut and paste” mechanism
Less than 2% of human genome composed of DNA transposons
Uses Terminal Inverted Repeats (TIR), Flanking Direct Repeats (DR)
Terminal Inverted Repeats (TIR)
Inverted compliments of 9-40 base pairs located at both ends of the transposable element. It is recognized by the transposase
Flanking Direct Repeats (DR)
Not a direct part of the transposable element but plays a role in inserting the transposable element back in the genome. It provides a marker for the excision site once a TE has moved. It is part of the DNA and dictates where the TE goes.
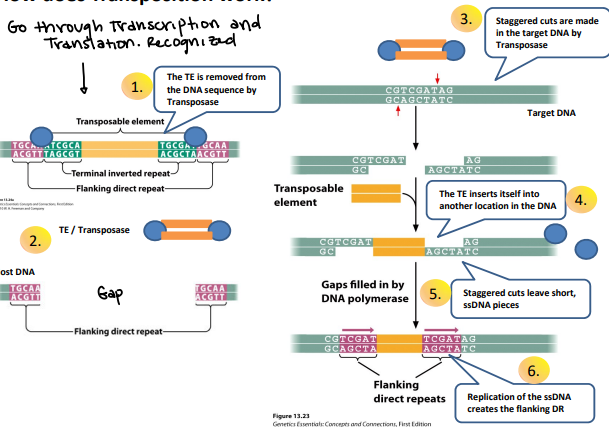
How does transposition work?
Before: Goes through transcription and translation, then it is recognized
The TE is removed from the DNA sequence by transposase
TE/Transposase
Staggered cuts are made in the target DNA by transposase, makes overhanging ends
The TE inserts itself into another location in the DNA
Staggered cuts leave short, ssDNA pieces
Replication of the ssDNA creates the flanking DR
Mechanism of Class 1 TEs
Functions through the action of RNA intermediates
Does NOT encode Transposase; encodes reverse transcriptase
Produces RNA transcripts and relies on reverse transcriptase to reverse transcribe the RNA into DNA sequences prior to insertion into target DNA
Moves through the genome in a “copy and paste” mechanism
Has Long Terminal Repeat transposons (LTR) and Non-Long Terminal Repeat transposons (NLTR)
Long Terminal Repeat (LTR) Transposons
Characterized by the presence of LTR on each end of the TE. Each end has satellites of repeated DNA and can be autonomous
Not in human genome
Non-Long Terminal Repeat (NLTR) Transposons
LTR is not present and is the ONLY active class of transposons in humans
Autonomous and non-autonomous
Autonomous (Long Interspersed Elements- LINEs)
Capable of moving on their own as they make their own Reverse Transcriptase required to move
DNA sequences that range in length from a few hundred to as many as 9,000 base pairs; they’re very long because they actually have the gene for reverse transcriptase and can make their own proteins.
Functional L1 elements are about 6,500 bps in length and encode proteins, including an endonuclease that cuts DNA and a reverse transcriptase that makes a DNA copy of an RNA transcript
Most L1 elements are not functional, they may play a role in regulating the efficiency of transcription of the gene in which they reside
Non-autonomous (Short Interspersed Elements- SINEs)
Must “borrow” reverse transcriptase from another element in order to move. Cannot make reverse transcriptase, so they use reverse transcriptase that comes from LINEs.
Short DNA sequences (100-400 bps) that represent reverse-transcribed RNA molecules originally transcribed by RNA pol II
Represents some 10% of total DNA
Most abundant SINEs are the Alu elements consisting of a sequence that contains a site that is recognized by the restriction enzyme Alu1
Alu elements
A transposon; it codes for all the proteins and enzymes needed to move a transposon
5’-AG/CT-3’ or 3’-TC/GA-5’
Occurring in the introns of genes which can be spliced into mature mRNA creating a new exon, which will be transcribed into a new protein product (way of possibly creating new proteins)
Alu1 Mechanism
If the Alu1 happens to sit down in the middle of the intron, then it is spliced along with the exons. Then the Alu1 will become an exon, if this will be able to code for a protein, then it is a new protein product. Increasing the genetic diversity.
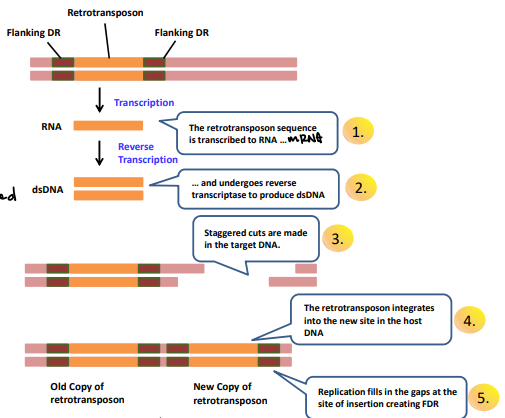
How does retrotransposition work?
Retrotransposon sequence is transcribed to RNA.. mRNA
Undergoes reverse transcriptase to produce dsDNA, winded up with double stranded molecule
Staggered cuts are made in the target DNA
Retrotransposon integrates into the new site in the host DNA
Replication fills in the gaps at the site of insertion creating FDR, unbalanced. Copied and added DNA
What do Transposable Elements do?
For the most part, it just depends on where it lands.
In non coding regions, nothing much
In genes, it can result in a mutation; however, keep in mind that not all mutations are deleterious
What leads to genetic diversity?
Mutations