Unit 9 The Cytoskeleton & Intro to Light Microscopy
1/138
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
139 Terms
light microscopy
extends human vision to resolve structures and kinematics at tissue-, cell-, and subcellular scales
fluorescence
physics; chemical vs genetically encoded proteins
filters
allow detection of emitted and separation from excitation light
cytoskeleton
a dynamic polymeric structure made possible by a myriad of protein-protein interactions:
Monomer subunit vs filament; covalent vs non-covalent bonds; single-filament vs bundles; polar vs nonpolar filaments; network morphology: branched, entangled, cross-linked…
filament
built from linear polymerized monomers that add noncovalently; move and attack → dynamic cytoskeleton
light microscopes
extend human vision in magnification (size), resolution (spatial), and contrast (signal-to-noise); Use wave- and particle-like properties of photons and how they interact with samples.
brightfield
light is absorbed
phase
light is phase shifted
differential interference contrast
light is differentially phase shifted
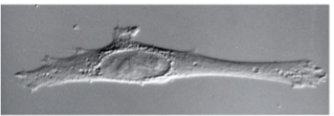
fluorescence microscopy
light is absorbed and re-emitted; enables to see where individual or specific abundances of proteins occur
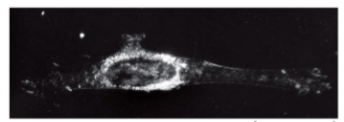
goal of using fluorescence
to understand molecular origins of cell biological processes:
When proteins associate with each other (polymerization)?
Where proteins are in the cell (polarity)?
How proteins move in a cell (diffusion, motors, stability, dynamics)?
Who proteins associate with in the cell (density, multiple fluors, protein complexes)?
fluorescence
the physical ability of a substance to absorb light of one color and emit light of another
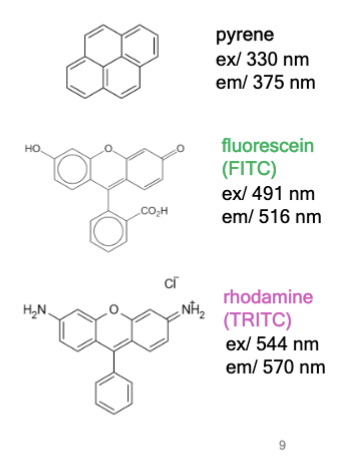
fluorophore
chemical compound that can absorb photons in the “ground state,” leading to fluorescence emission and vibration relaxation in the “excited state.”
examples: pyrene, fluorescein, and rhodamine
fluorescent proteins
genetically encoded proteins capable of producing fluorescence through the spontaneous formation of fluorophores from certain amino acid residues, allowing for the visualization of specific molecular processes, organelles, and organisms in their native conditions.
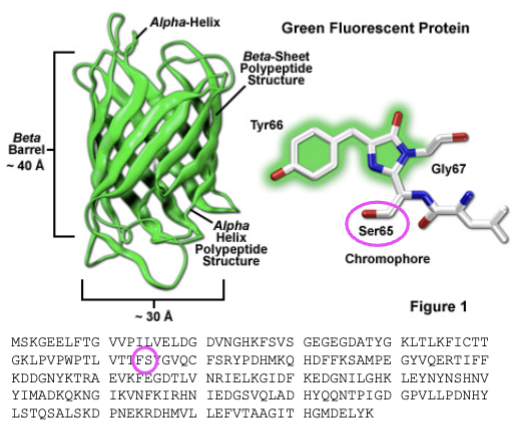
green fluorescent protein
first FP discovered; discovered in jellyfish; absorbs blue light and emits green;
what makes fluorescent proteins special
they can be genetically encoded in a genome; combined with any other gene domain; targeted to any cell, any tissue, any time
mutations in green fluorescent protein
S65T and F64L — introduced _______ that improved spectral properties and protein folding; able to be better synthesized in humans
wavelength
Fluorescent materials absorb light of one _________ and emit light at a lower energy _________.
Stokes shift
Difference in wavelengths; some energy is dissipated as heat when a photon is emitted, leading to this from excitation to emission
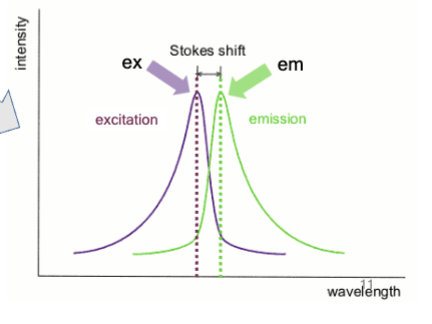
excitation
how much the fluorescent material absorbs of a particular wavelength
emission
emitting a photon
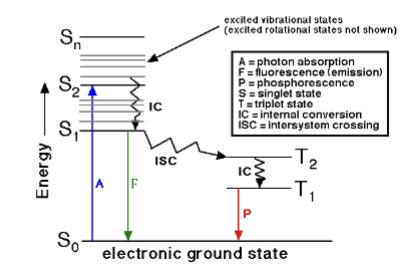
Jablonski Diagram
explains absorption and emission spectra of fluorescent materials.
Electrons at high energy orbital states fall down in small jumps, and then the emission is a large jump which is the fluorescence you see
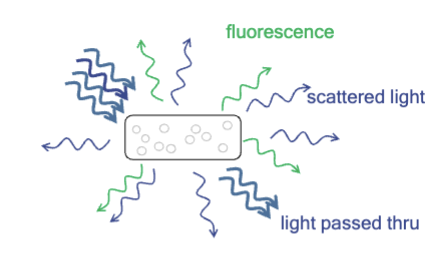
problem with fluorescence
How do you distinguish excitation light that is reflected or scattered from the sample from emitted light?
How to tell what is what is noise and signal?
propagation of light
Control the ___________ through the optical train with barrier and beam-splitting mirrors.
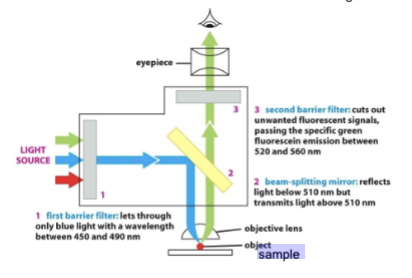
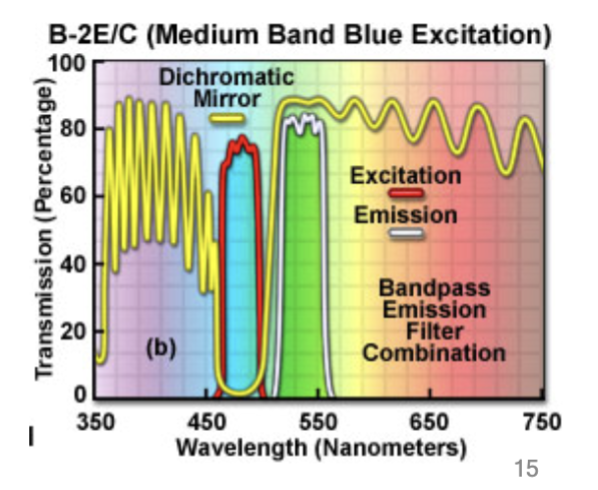
barrier filter
#1: When dealing with fluorescence, use this to only allow precise wavelengths of light for excitation. This first lets through only blue light with a wavelength between 450 and 490 nm.
beam-splitting filter
#2: Place this in the optical train that reflects excitation but passes emission wavelengths. This reflects light below 510 nm but transmits light above 510 nm
block unwanted
Place an additional a barrier filter (#3) to __________ wavelengths. (Because #1 and #2 filters are not perfect); cuts out [____] fluorescent signals, passing the specific green fluorescein emission between 520 and 560 nm
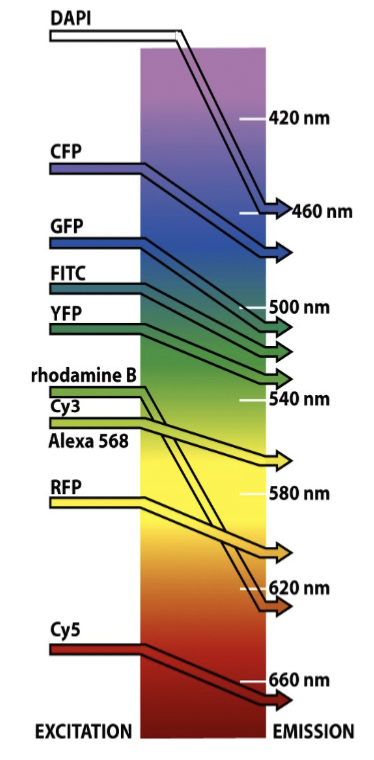
fluorophores
molecules or proteins (e.g. fluorescent proteins) that absorb at one wavelength and emit at another; can control the excitation wavelength by choosing a light source, such as mercury, halogen, LED, or a laser.
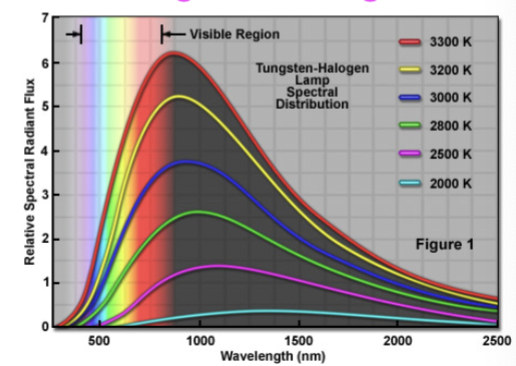
Tungsten Halogen
broad spectrum light source
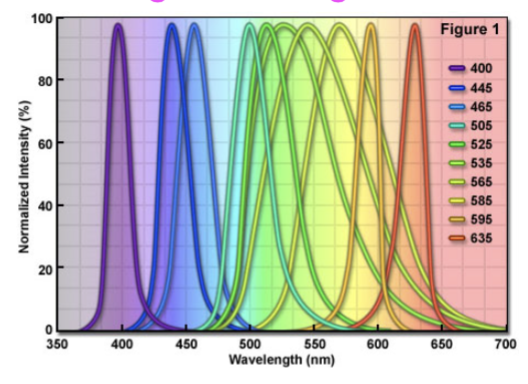
light emitting diodes
aka LEDs; narrow wavelengths of light
scattered excitation light
in fluorescence microscopy, you must be able to separate light emitted by fluorophores from the background of this.
filters
based on the spectrum of the fluorophore, you need to construct a set of …
excitation or emission
barrier or band-pass filters
beam-splitter
dichromatic mirror - reflects excitation and passes emission
histology fixation
when live imaging is not practice or possible (in vivo samples from animals or patients) use this; involves chemical cross-linking with formaldehyde or glutaraldehyde, or precipitation with alcohols (ethanol, methanol), or precipitation with acids
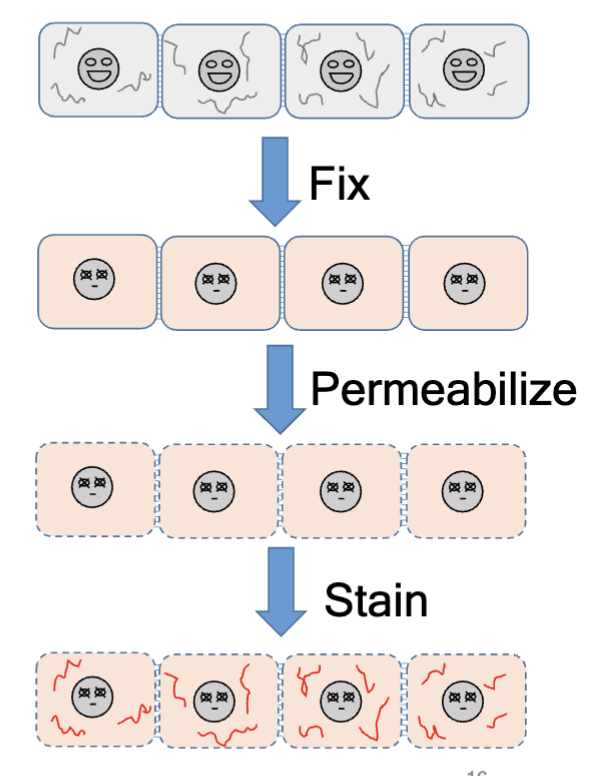
permeabilization
after fixation, do this with Triton X-100, Tw-20, etc; create holes in the plasma membranes
stain
after permeabilization, _____ specific structures or complexes using immunostaining, DNA (DAPI or Hoecsht), phalloidin (F-actin); see proteins in localized positions (where they were when they were fixed)
immunostaining
“see” specific molecules using this; To improve contrast we can place absorbing and fluorescent molecules (markers) using antibodies raised to specific target molecules and proteins
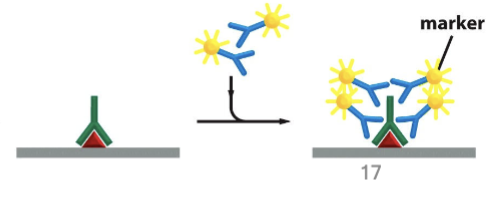
markers on antibodies
can be…
enzymes such as peroxidase that will deposit absorbent particles visible in brightfield
radioactively labeled molecules that will emit radioactivity that can be detected
fluorescent molecules
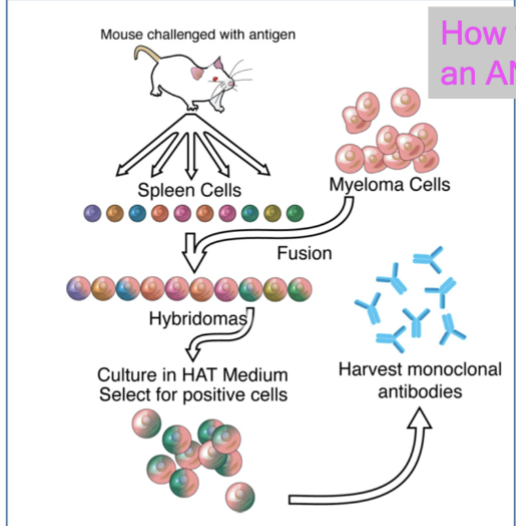
how to make an antibody
mouse challenged with antigen → collect spleen cells → add myeloma cells → form hybridomas → culture in HAT medium, select for positive cells → harvest monoclonal antibodies
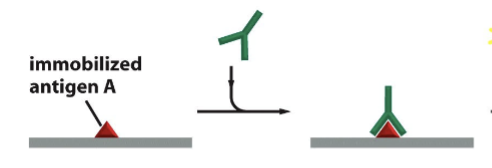
primary antibody
rabbit antibody directed against antigen A
secondary antibodies
marker-coupled antibodies directed against rabbit antibodies
load reporter substances
the major challenge for live cell microscopy is that you need to _______________ into cells via microinjection, electroporation, lipofection, gene gun, or genetically encoded reporters
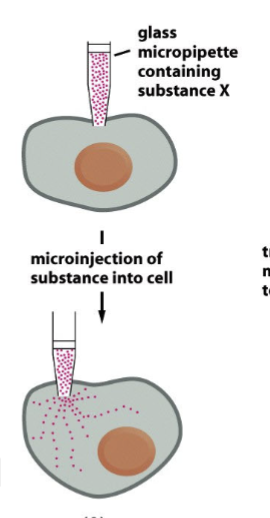
microinjection
glass micropipette containing substance X against the plasma membrane → _______ of substance into cell
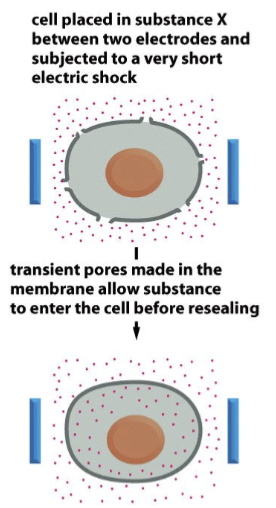
electroporation
cell placed in substance X between two electrodes and subjected to a very short electric shock; transient pores made in the membrane allow substance to enter the cell before resealing.
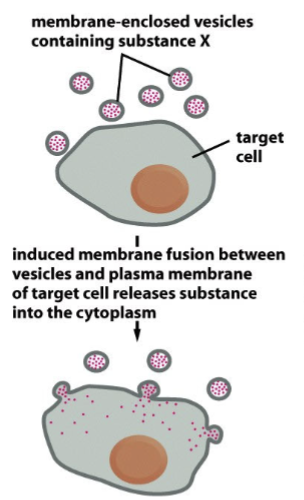
lipofection
membrane-enclosed vesicles containing substance X move towards target cells; induced membrane fusion between vesicles and plasma membrane of target cell releases substance into the cytoplasm
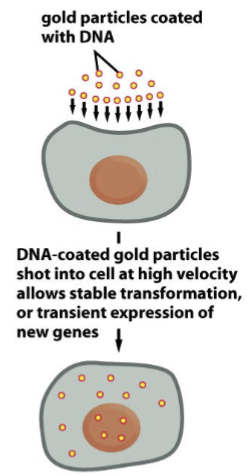
gene gun
gold particles coated with DNA; and then these DNA-coated gold particles are shot into the cell at high velocity, which allows stable transformation or transient expression of new genes
molecular complexes
carry out every key task in the cell
cytoskeleton components
ribosomes, thick microtubules, F-actin filaments, intermediate filaments; all provide surface area inside the cell for proteins to be localized to

affinity chromatography
“fishing” for target proteins; use “bait” that will allow you to capture your target; how can we find, identify, and test the involvement of specific molecules in these tasks.
essentially use a refined bait to catch a lot of proteins and purify them
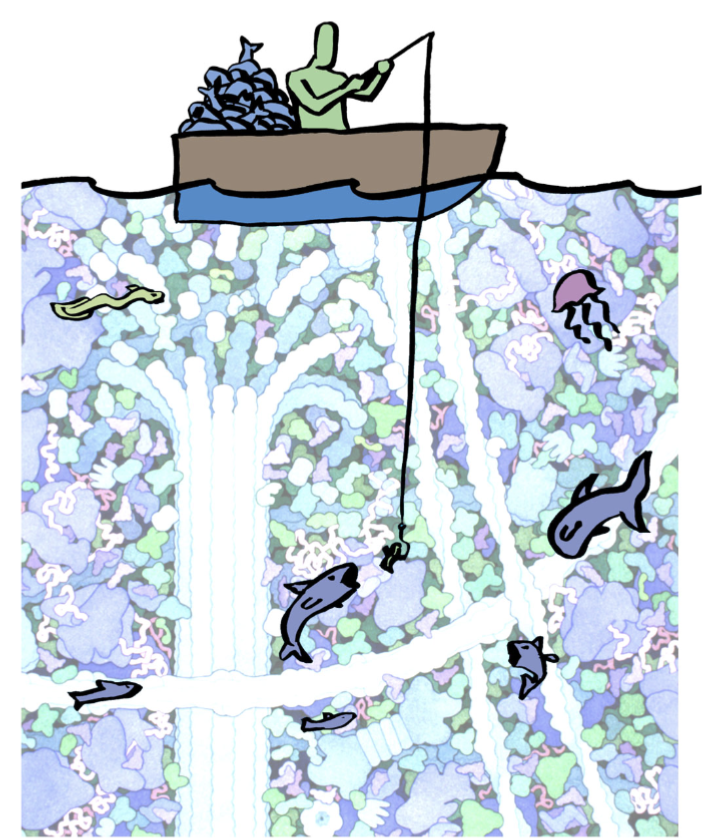
experimental technique: affinity chromatography
Used to separate and purify both known and unknown factors from cells and/or tissues. Relies on non-covalent interactions between the factor and a known binding partner
example of affinity chromatography with f-actin
Seeking proteins in cytoplasm that bind f-actin
To set up your column:
purify f-actin and stabilize with phalloidin
add a biotin group to f-actin (covalently modify)
bind F-actin to streptavidin and form strong (!!!!) non-covalent bonds
“pack the column” - place beads into the glass column that retains the beads but passes solutions
pour cell lysate through the column; critical “ocean to fish in” - whatever in lysate that likes to bind to actin binds
Wash the column with wash buffer that elutes non-binding proteins
Elute "hypothetical" proteins from the column - use a buffer that disrupts protein-protein interactions.
Then, ASSAY your proteins! (immunofluorescence, reporter, pyrene actin, TIRF, etc)
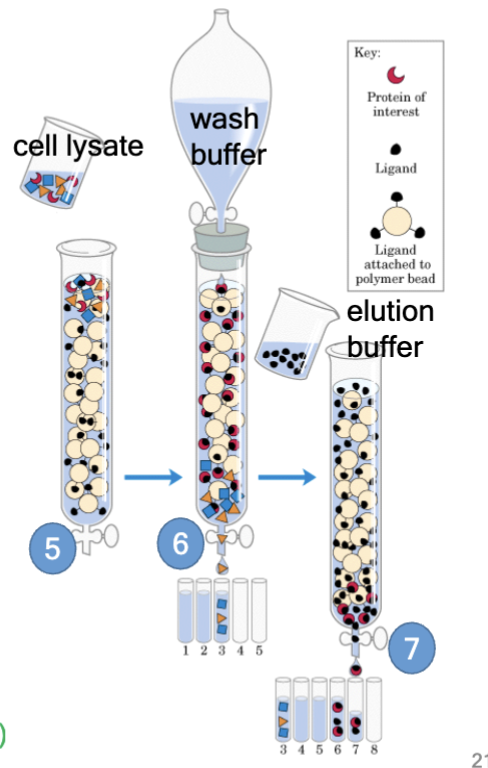
protein of interest
in affinity chromatography, the “hypothetical” protein you seek
ligand
in affinity chromatography, the protein you already have (bait)
ligand attached to polymer bead
in affinity chromatography, a way to retain hypothetical proteins in the column
in columns or in bulk
Affinity Chromotography ___________ have been used to find and purify many actin-binding or microtubule-associated proteins
after affinity chromatography
Further analysis of proteins collected from this technique:
1) Size – from Western Blots: PAGE.
2) Complex members – Immunoprecipitation then Western Blot.
3) Amino-acid sequence - from Mass Spectrometry.
4) Kinetics of protein-protein interaction - from Surface Plasmon Resonance.
antibodies
make great “bait”
other baits
proteins: fragments, whole proteins, or complexes
find and purify
Affinity Chromotography commonly used to _______ many protein complexes including actin-binding, microtubule-associated proteins, and intermediate filament associated proteins.
size
Further analysis of proteins collected from affinity chromatography to find this from Western Blots. (antibodies to detect)
additional members of protein complex
further analysis of proteins collected from affinity chromatography to find this via Immunoprecipitation then Western Blot. (antibodies to precipitate)
identify sequence
further analysis of proteins collected from affinity chromatography to find this via Mass Spectrometry.
Kinetics of protein-protein interaction
further analysis of proteins collected from affinity chromatography to find this via multiple approaches after making a biosensor (protein::FP) including TIRF, live-cell imaging, FRAP
cytoskeleton modulators
specific application of affinity chromatography is to find and test the role of __________; use actin subunits or filaments as “bait” bound to the column
actin
the form and function of this in the cell is controlled by a diverse set of proteins that control how filaments form and associate with one another
each subunit has polarity, and there is a ATP binding site that binds ADP when in the filament
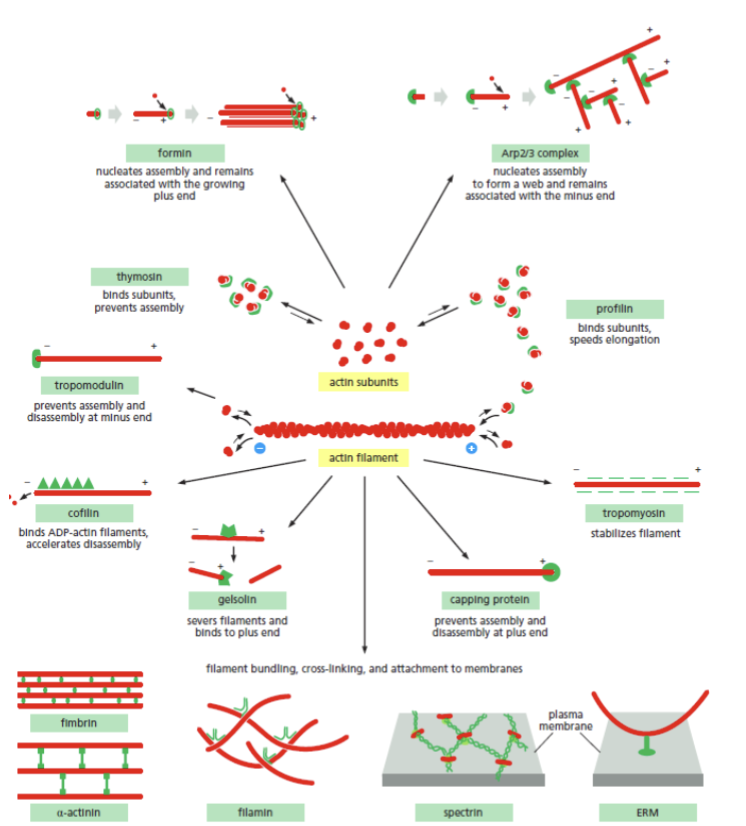
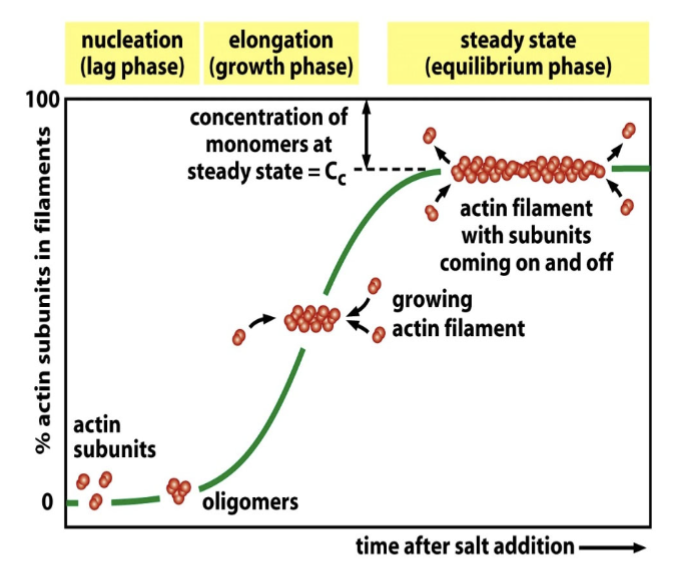
actin polymerization
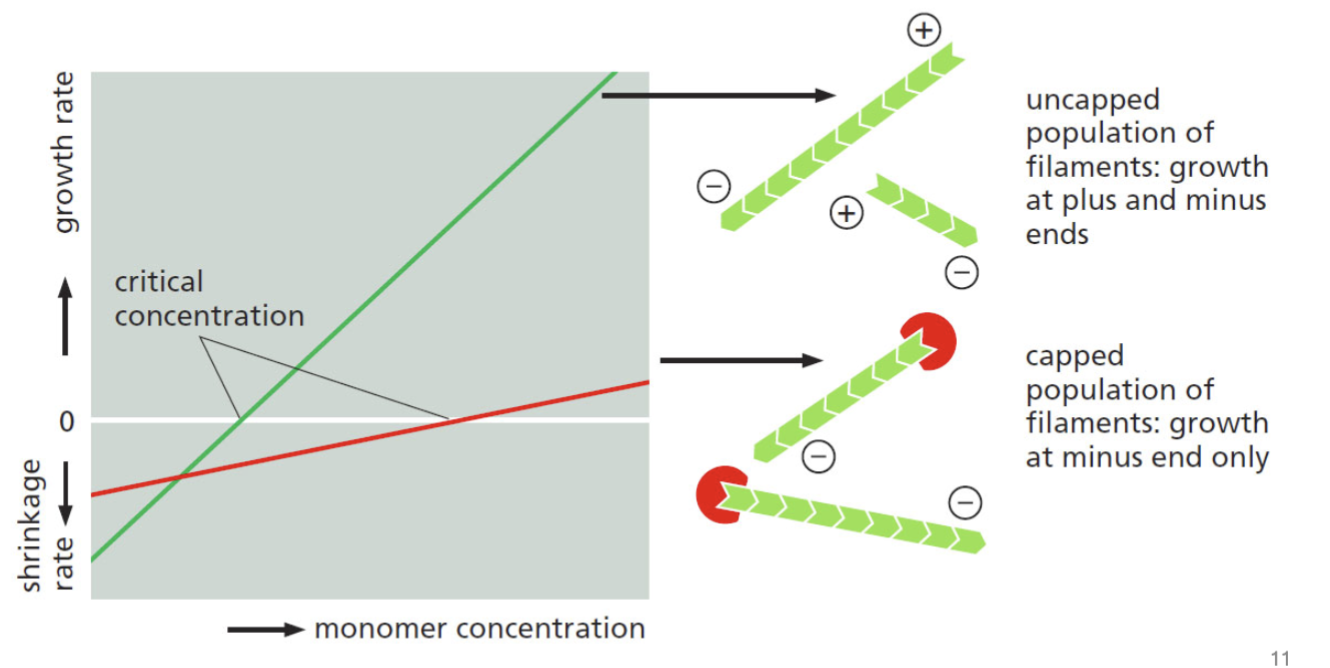
highly regulated and non-covalent; occurs at positive end, while the negative end is cut back; F-actin has this regulated by G-actin binding to ADP or ATP. Each end of the filament has different critical concentration
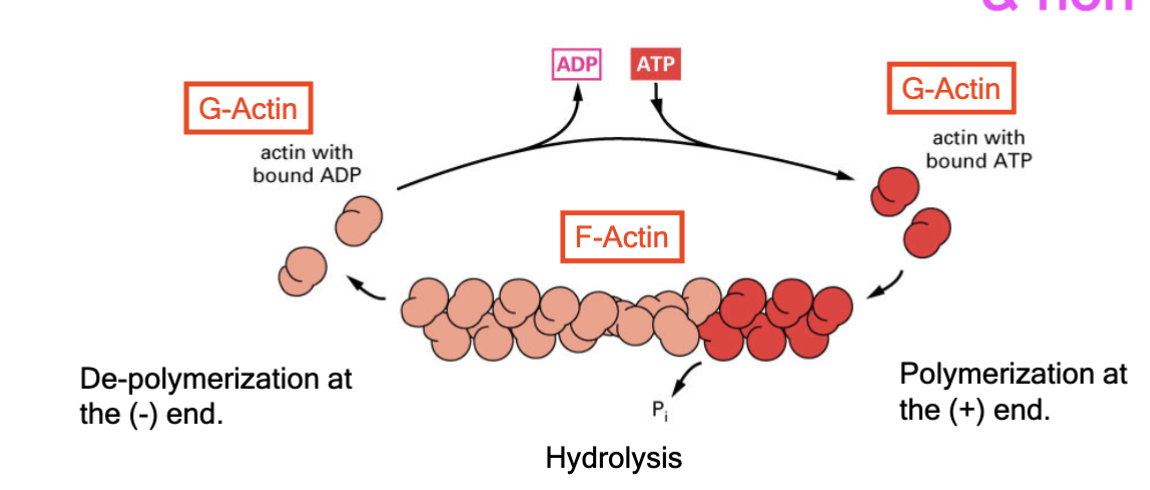
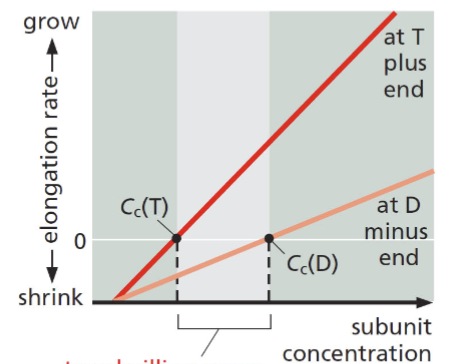
treadmilling
for F-actin and microtubules, a consequence of nucleotide hydrolysis that accompanies polymer formation is to change the critical concentration at the two ends of the polymer. Cc (minus end) > Cc (plus end). If both ends of a polymer are exposed, polymerization proceeds until the [ ] of the free monomer reaches a value that is above Cc for + and below Cc for —. At this steady state, subunits undergo a net assembly at the + end and a net disassembly at the — end at an identical rate. Polymer maintains constant length, even though there is a net flux of subunits through the polymer
![<p>for F-actin and microtubules, a consequence of nucleotide hydrolysis that accompanies polymer formation is to change the critical concentration at the two ends of the polymer. C<sub>c</sub> (minus end) > C<sub>c</sub> (plus end). If both ends of a polymer are exposed, polymerization proceeds until the [ ] of the free monomer reaches a value that is above C<sub>c</sub> for + and below C<sub>c</sub> for —. At this steady state, subunits undergo a net assembly at the + end and a net disassembly at the — end at an identical rate. Polymer maintains constant length, even though there is a net flux of subunits through the polymer</p>](https://knowt-user-attachments.s3.amazonaws.com/a427a582-7653-4b04-adec-476c99dd04e9.png)
pyrene actin
This chemical is more fluorescent (glows when excited with specific wavelengths of light) when it is in a filament
pyrene actin assay
Function of actin binding proteins also explored with other biochemical methods such as immunoprecipitation, immuno-localization, live imaging, and TIRF microscopy.
1) Set up a reaction including the chemical monomer in a transparent vessel (cuvette) with or without “factor X”
2) Follow intensity of emitted fluorescence
3) Compare time of different phases, slopes, equilibrium, rates of change in the growth phase
fixed
at the start of the pyrene actin assay reaction, the number of subunits in the cuvette is….
decreases; increases
as the pyrene actin assay reaction proceeds, the number of free subunits _______ and the amount of polymer _________
polymerization chemistry
pyrene actin assay quantifies this; the time course is that the assembly of a protein into a long helical polymer such as a cytoskeletal filament or bacterial flagellum typically shows the following time course
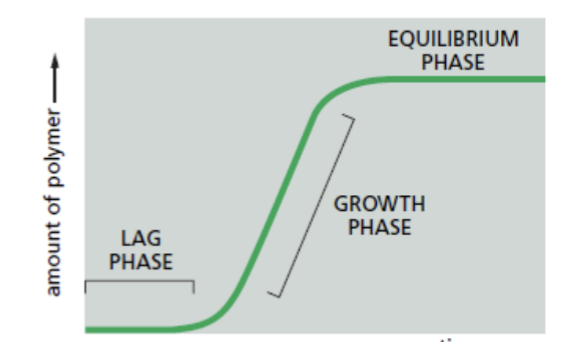
lag phase
corresponds to time taken for nucleation
growth phase
occurs as monomers add to the exposed ends of the growing filament, causing filament elongation
spiked with high [G-actin] → higher [subunits] → higher number of net amount of F-actin
equilibrium phase
steady state → reached when the growth of the polymer due to monomer addition precisely balances the shrinkage of the polymer due to disassembly back to monomers
variables in pyrene actin assay
concentration at start; specific proteins; change from steady state
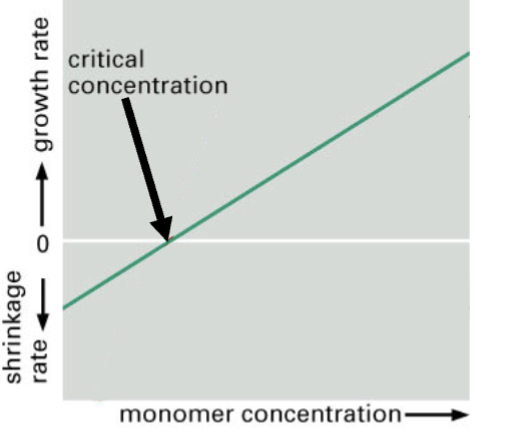
monomer concentration vs rate of change of amount of polymer
a different way to express graphically the results of a pyrene actin assay;
+ rate → + and - ends growing, starting at the middle point of that + line
at the net critical concentration, depleting the monomer and it is not possible to have the same number of monomers coming off the negative ends as going onto the positive end
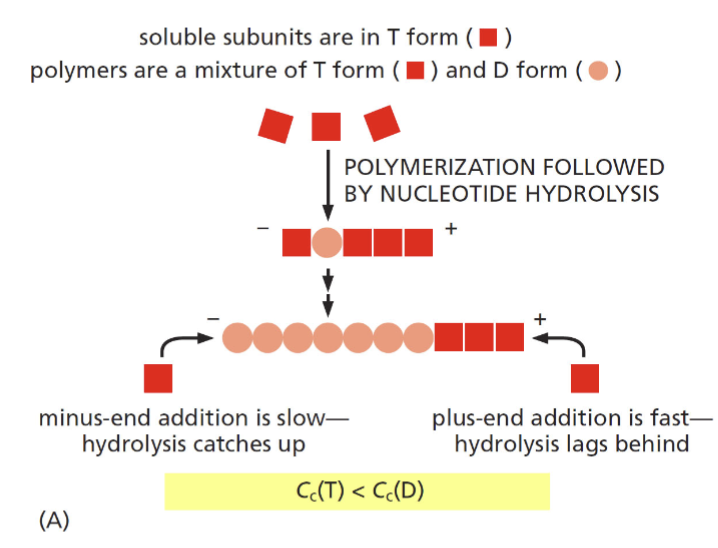
ATP hydrolysis and F-actin structure
two factors that lead to different rates of polymerization at the (+) and (-) end
hydrolysis at the minus end catches up as addition is slow (requires high abundance of monomers to add); hydroylsis lags behind and plus end as addition is fast
treadmilling range
get to the same amount of polymerization at the positive end as depolymerization at the negative end (equilibrium)
gelsolin
cleaves actin filaments and regulates polymerization by increasing the number of positive ends; present in the cytosol of the bloodstream; meant to prevent actin from clogging up the cardiovascular system. Caps the filament
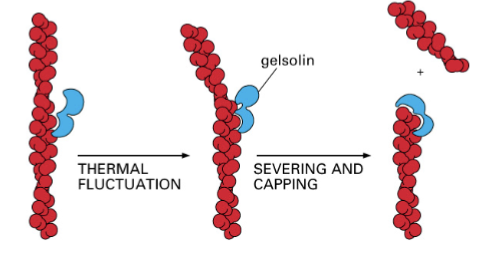
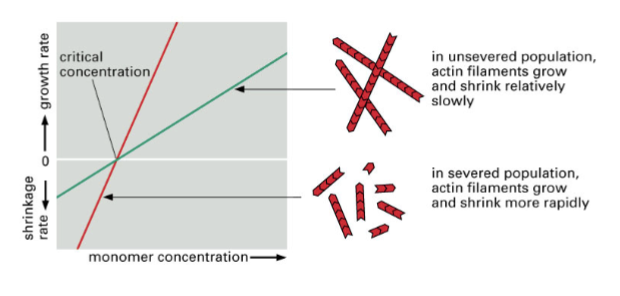
faster rate of depolymerization
gelsolin changes the ratio of polymerization so that it occurs more at the negative end, meaning this…
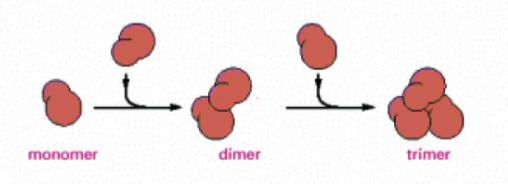
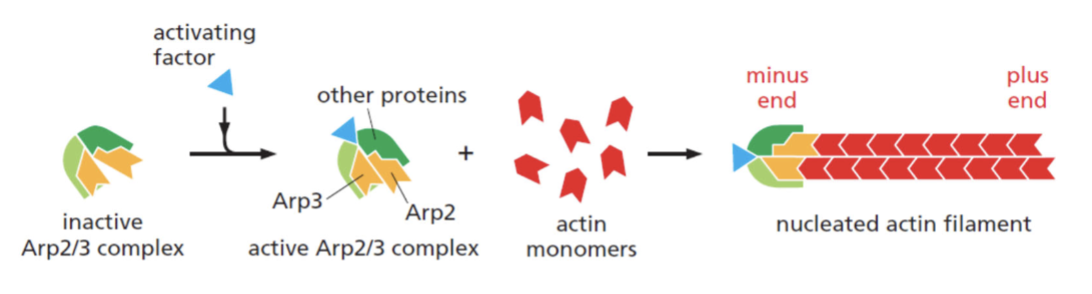
nucleation
helical polymer stabilized by multiple contacts between adjacent subunits. Two actin molecules bind relatively weakly to each other, but adding a third to form a trimer makes the entire group more stable. Further monomer addition can take place onto this trimer, which acts as a nucleus for polymerization. For tubulin, nucleus is larger and has a more complicated structure (same principle).
This process is relatively slow, explains lag phase during polymerization
slowed or eliminated
the lag phase can be ____________ entirely if premade nuclei, such as fragments of already polymerized microtubules or actin filaments, are added.
Example: Arp2/3 complex allows immediate polymerization without a nucleation step
plus-end capping protein
can alter polymerization rates; heterodimer that binds to the plus end of actin filaments, preventing subunit exchange at this site. No sites for G-actin to add easily, and the only available site (minus end) binds poorly to G-actin
Biochemistry
Affinity chromatography.
Proximity-dependent biotinylation.
Immunolocalization.
Immunoprecipitation.
Western blot.
How much is in a cell?
What are the binding partners?
polymerization assay
Pyrene actin assay
What are the effects of binding partners on nucleation and polymerization rates?
Total Internal Reflection Fluorescence Microscopy (TIRF)
How do binding partners alter the F-actin network? For instance: actin elongation, severing, and branching assay
molecular-scale function
Biophysical studies provide information about this:
1) Molecular function (rates, step-size…)
2) Biochemistry (ATPases rates,…)
3) Mechanical function (force-velocity,…
choosing LED
chose below the excitation peak of the compound for best results
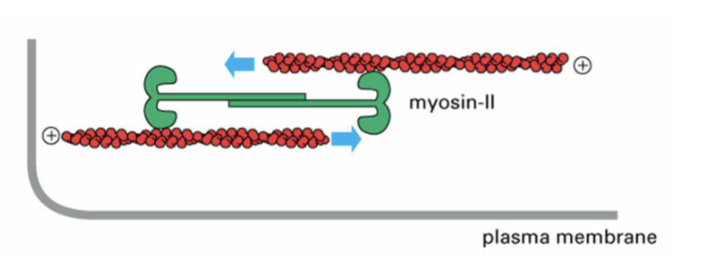
myosins
motors that step along F-actin; first motor protein was skeletal muscle version of this; are ATPases
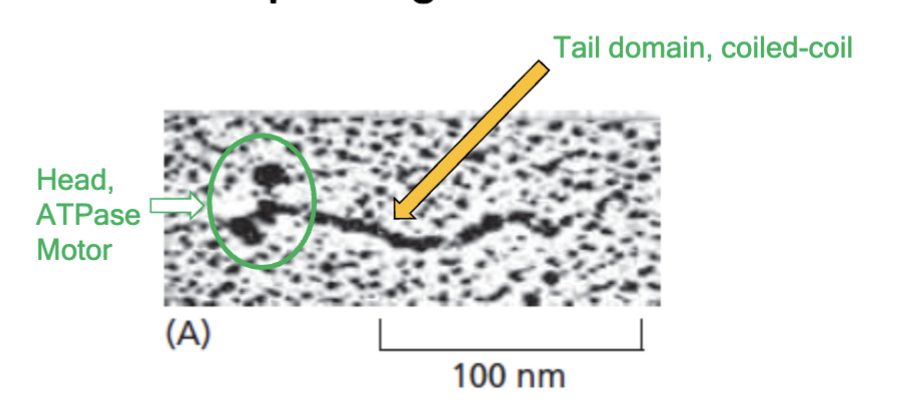
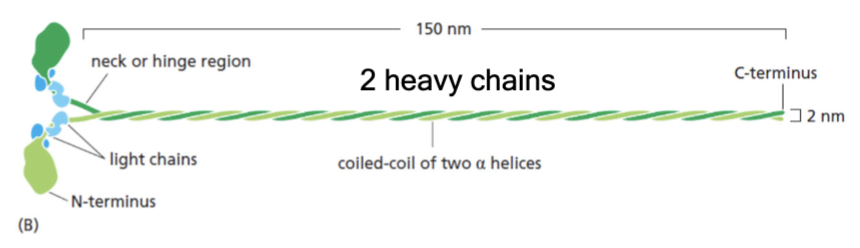
structure of myosin
two heavy chains; four light chains; force-generating globular head; long coiled-coil tail
motors
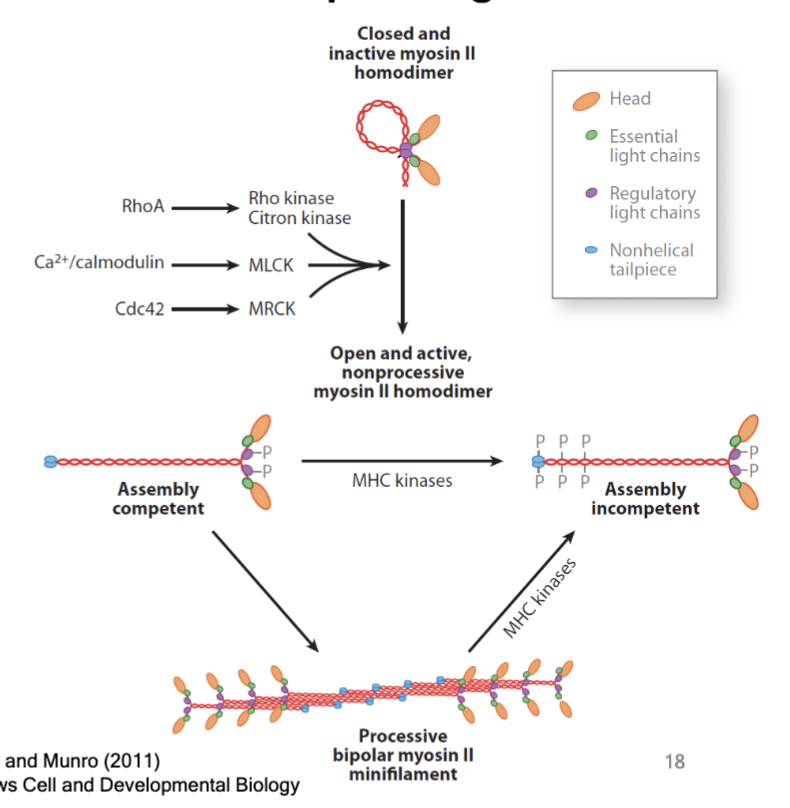
can be regulated like other proteins; kinases phosphorylate the light chain and heavy chain in myosin.
phosphorylation of light and heavy chains
control enzymatic activity of the myosin and control assembly into minifilament “working structure”
mechanical energy
motors convert chemical energy to _____________.
steps to convert chemical to mechanical energy
1) Myosin II binds F-actin
2) ATP binds myosins II, causes release from F-actin
3) Hydrolysis of ATP to ADP drives conformational change in myosin II head - moves in “forward” direction (phosphate transiently attached to myosin)
4) De-phosphorylation (loss of phosphate) allows myosin II to bind F-actin
5) Release of ADP allows myosin II power stroke: myosin II and F-actin move “backward”
[Repeat]
![<p>1) Myosin II binds F-actin</p><p>2) ATP binds myosins II, causes release from F-actin</p><p>3) Hydrolysis of ATP to ADP drives conformational change in myosin II head - moves in “forward” direction (phosphate transiently attached to myosin)</p><p>4) De-phosphorylation (loss of phosphate) allows myosin II to bind F-actin</p><p>5) Release of ADP allows myosin II power stroke: myosin II and F-actin move “backward”</p><p>[Repeat]</p>](https://knowt-user-attachments.s3.amazonaws.com/fd8840de-f62a-47e7-b65c-20aad369ce53.png)
distinctive tail structures
there is a diverse myosin gene family that has monomer or dimer of motors with this. Myosin ATPase is in the motor domain.
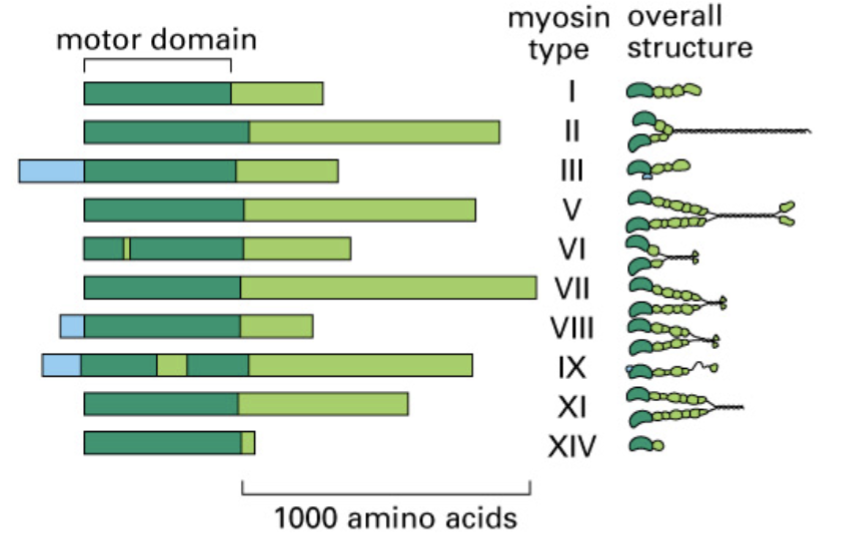
functions dictated by structure
true for myosin isoforms.
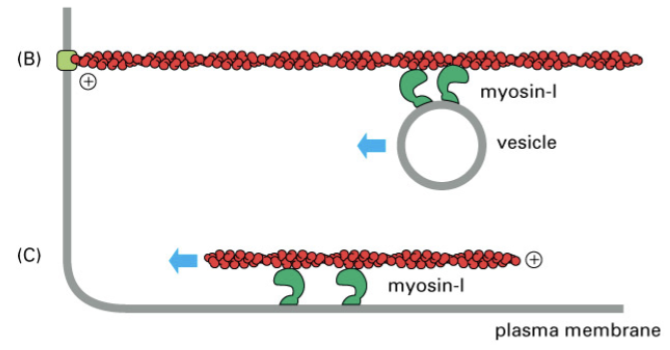
myosin I
has only one head and one tail but works coordinately to transport vesicles or F-actin
myosin II
forms multiple oligomers of dimers, forming mini thick filaments, to reorganize F-actin filaments