AP bio unit 6 genetics
1/99
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
100 Terms
central dogma
the flow of genetic information in cells, from DNA to RNA, then from RNA to polypeptide (DNA replication, transcription, translation)
retroviruses
viruses that have an RNA genome (instead of a DNA genome), including an enzyme (reverse transcriptase) that allows it to go through REVERSE transcription, using its RNA to synthesize DNA
reverse transcription
rna is used as a template, pairs up with one DNA strand, then DNA replicates to create 2 strands (eg. in HIV, a retrovirus).
nucleus
where does (DNA) replication occur in eukaryotes?
nuceloid
where does (DNA) replication occur in prokaryotes?
multiple linear (multiple strands)
structure of DNA in eukaryotes?
single circular (it literally only has 1 DNA (still double helix))
structure of DNA in prokaryotes?
a, t, c, g
what nitrogenous bases is DNA made up of?
pentose sugars (deoxyribose), and phosphate groups
what is the backbone of DNA made of?
nucleotide
the combination of a pentose sugar, phosphate group, and nitrogenous base
purine
has a double ring structure, includes bases A and G
pyrimidine
has a single ring structure, includes bases C and T (and U in RNA)
2 (hydrogen bonds)
how many bonds do A and T have?
3 (hydrogen bonds)
how many bonds do G and C have?
5’ end
which end of the DNA has the phosphate?
3’ end
which end of the DNA has the hydroxyl group?
yes (think of two lane traffic, cars go opposite each other on different lanes)
are DNA strands antiparallel?
3 to 5
what way is DNA READ?
5 to 3
what way is DNA SYNTHESIZED?
DNA helicase
unwinds the DNA strands by breaking the hydrogen bonds between bases, 1st step in replication
topoisomerase
relaxes supercoiling in FRONT of the replication fork (think when untwisting two strands twisted together, when you pull one side to untwist, the other side gets tighter, or supercoils that’s how it made sense to me)
replication fork
where the DNA separates during replication, in the shape of this utensil
primase
synthesizes the RNA primer (DNA polymerase requires RNA primers to initiate DNA synthesis)
RNA primer
a series of nitrogenous bases (AGCU) created by primase to tell DNA polymerase where to start replication; DNA polymerase goes back and removes/edits these later
DNA polymerase
synthesizes new strands of DNA continuously on the leading strand and discontinuously on the lagging strand
leading strand
synthesized continuously TOWARDS the replication fork (the ‘3 to ‘5 strand)
lagging strand
synthesized discontinuously AWAY from replication fork ( the 5’ to 3’ strand), since DNA polymerase reads 3’ to 5’, it has to keep doubling back and reading/synthesizing as more DNA is unraveled (this needs multiple primers)
ligase
joins the fragments (okazaki fragments) on the lagging strand; “seals the bond” between fragments by joining the 3’ and 5’ ends
okazaki fragments
fragments of DNA synthesized on the lagging strand because DNA polymerase has to keep doubling back
SSBs
proteins that keep the separated single strands from coming/joining back together
DNA replication
a semi-conservative process (keeps the old strands but adds new strands through replication) that makes new DNA
transcription
process of turning DNA into mRNA
nucleus
where does transcription occur in eukaryotes?
nucleoid (cytosol)
where does transcription occur in prokaryotes?
a, u, c, g
what nitrogenous bases is RNA made of?
pentose sugars (ribose), and phosphate groups
what makes up the backbone of RNA?
3 to 5
what way is mRNA READ?
5 to 3
what way is mRNA SYNTHESIZED?
template strand
the 3’ to 5’ strand, the OPPOSITE of its bases make the mRNA strand
coding strand
the 5 to 3 strand, its bases are the exact same as the mRNA strand (except for t replaced with u)
RNA polymerase
separates the DNA strand and synthesizes mRNA molecules in the 5 to 3 direction by reading the template DNA strand in the 3 to 5 direction
promoter
the site where RNA polymerase binds to start transcription (made of series of nitrogenous bases)- just a region
transcription factors
activators/inhibitors that turn on/off gene expression and binds to the DNA, affecting the ability of RNA polymerase to bind
eukaryotes
are post-transcriptional modifications present in eukaryotes or prokaryotes?
post transcriptional modifications
changes that occur to RNA transcripts after they are transcribed from DNA, but before they are translated into proteins, and include processes like capping, splicing, and polyadenylation.
5’ guanine cap (capping)
signals the “start” of the mRNA transcript for ribosome to bind, facilitates export from nucleus, allows mRNA to come out of the nucleus into where the ribosomes are (cytosol)
splicing
removal of introns from pre-mRNA transcript, cuts out unnecessary sequences/non-coding regions
introns
unnecessary sequences and non-coding regions cut out during splicing (post transcriptional/mRNA modification) in eukaryotes
poly a tail
a bunch of adenines put at the end of the mRNA (3’ end), inhibits DEGRADATION from hydrolytic enzymes in cytosol and prolongs the “life” of mRNA
translation
going from mRNA (RNA) to polypeptide (from one language to another)
NO
do prokaryotes cut out introns?
rRNA (ribosomal RNA)
RNA that makes up ribosomes
large subunit
the subunit of a ribosome that binds tRNA (transfer RNA)
tRNA (transfer RNA)
carries a specific amino acid to the ribosome, to correctly translate from mRNA to polypeptide
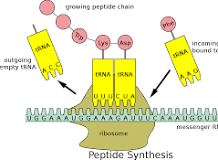
small subunit
the subunit of a ribosome that binds mRNA (messenger RNA)
mRNA (messenger RNA)
the RNA that is synthesized from DNA, and brings the genetic “message”/code to correctly code for polypeptides
cytosol and rough ER
where does translation occur in eukaryotes?
cytosol
where does translation occur in prokaryotes?
True
T/F: all ribosomes start out in the cytosol, then receive a message to attach to the rough ER
initiation, elongation, termination
what are the 3 steps of translation?
initiation
start codon “activated” (AUG), methionine created
AUG, methionine
what is the start codon and amino acid for all proteins?
elongation
the polypeptide is “elongated”, base pairs between tRNA and mRNA are attached, and amino acids attached with the tRNA to the growing chain.
termination
the ribosome hits the “stop” codon, and stops making the polypeptide
UAG, UAA, UGA
what are the 3 stop codons for translation?
a site
the site on a ribosome where the amino acid is added in, tRNA comes up to the mRNA and adds the amino acid and attaches the anti-codon which is going to base pairs up with the codon in mRNA
p site
where completed a site attachments (amino acids/tRNA) shifts over, where we have our growing polypeptide chain (this site holds the polypeptide chain together)
e site
an empty space for amino acids ready to exit on the ribosome, final shift/translocation in the ribosome
hydrolysis (signals for a water molecule to come in and break the polypeptide chain)
what happens when the ribosome reaches the stop codon?
yes
can DNA polymerase proofread/check itself?
nuclease (ligase then seals back together)
what does DNA polymerase to use to cut out mismatching bases to fix them?
NO (instead a release factor comes in which signals for water to come in and hydrolyze)
when the ribosome hits the stop codon, does it attach another amino acid?
changes it to DNA
what does DNA polymerase do to the RNA primer?
point mutations
mutations at ONE nucleotide base pairs (UAA→ UCA)
silent mutation
a type of point mutation that doesn’t change the amino acid once translated
missense mutation
a point mutation that changes from one amino acid to another when translated (UUA→GUA) (Leu→Val)
nonsense mutation
a point mutation that results in a premature stop (UUA→UAA) “don’t know the end of the sentence"
frameshift mutation
caused by the insertion or deletion of 1 or 2 nucleotide base pairs- shifts the “reading frame” for codons (UUA→UUGA, now the amino acids will have to group differently)
chromosomal mutation
rearrangement of chromosome parts or changes in chromosome numbers
insertion, deletion, duplication, inversion, translocation
types of chromosome rearrangements possible for a mutation?
nondisjunction, polyploidy
how can you have changes in chromosome number?
polyploidy
when you have a WHOLE nother set of chromosomes (you normally have 2, now you have 3)
operons
made up of a promoter, operator, and genes, which can be turned “on and off” to regulate gene expression and RNA synthesis
promoter
site where RNA polymerase binds
operator
site where repressor binds
repressor
STOPS RNA polymerase/Transcription → “rock on the train track”, does this by INHIBITING RNA POLY. FROM BINDING
repressible operons (this whole process happens in prokaryotes, not eukaryotes, although eukaryotes have a lot of the same parts)
these operons start ON, and a repressor attaching makes them INACTIVE (turned OFF); they usually synthesize something (some sort of anabolic pathways), but when they don’t need anymore, a repressor binds to halt RNA synthesis. EX: Trp operon, synthesizes tryptophan, if there’s too much tryptophan, it will act as a ligand and bind to the repressor to turn it OFF
inducible operons (this whole process happens in prokaryotes, not eukaryotes, although eukaryotes have a lot of the same parts)
these operons start OFF, and a repressor attaching makes them ACTIVE (turned ON); they usually break something down (catabolic pathway) In these cases, the repressor is always bound to the operon, but when a molecule needed to be broken down is present, it attaches to the repressor and the repressor FALLS OFF- ex: lac operon, synthesizes enzymes to break down lactose, always repressed, but if lactose is present, it binds to repressor to inactivate (the repressor), repressor falls off and the operon is turned ON
NEGATIVE
what type of feedback loop are both inducible and repressible operons?
CAP
a positive feedback loop, this is here if we want RNA polymerase to bind more often/effectively and produce more mRNA than it is already, and activated by binding to cAMP, that signals when glucose levels are low and we NEED MORE ATP (cAMP is formed through hydrolyzation of ATP)
gel electrophoresis
separating molecules based on size and charge- power is turned on and DNA fragments migrate though gel, fragments separated by size bc the smaller the fragment, the more it can travel through little holes in the gel and vice versa for larger fragments; thicker band=more DNA there, thinner band=less DNA
PCR (polymerase chain reaction)
makes multiple copies of DNA fragments, done if you’re trying to amplify your DNA
heating, cooling, annealing
steps of PCR in order?
heating
denatures the DNA and pulls it apart into single strands, 1st step in PCR
cooling
allows primers to bind to the specific places we want on the structure, 2nd step in PCR
annealing
this is when the DNA polymerase goes through and synthesizes DNA, 2rd step in PCR
TAQ polymerase
what polymerase do you have to use in PCR because of so many heating and cooling cycles?
bacterial transformation
introduces genetic material (plasmid) to bacteria, put plasmid into solution with bacteria, then heat-shock it to open up the membrane to allow plasmid to come in, then see which of them grow (if they have antibiotic resistance to the plasmid, they’ll still grow in the presence of the antibiotic)
DNA sequencing
using radioactive nucleotides to determine the sequence of a DNA strand, coding each of the different nucleotides
kind of (because they are separated and independently regulated, each with their own promoter regulating multiple genes, ex: the trp operon in eukaryotes controls 3 genes, and synthesizes the mRNA all at once (this is really confusing to me too))
do eukaryotes have operons?