Lecture #12 | Oncogenes in Signal Transduction: GF Receptors, Ras, Raf, MAPK, JNK, PKC
1/32
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
33 Terms
How was human Ras oncogene isolated
Isolated from human bladder carcinoma using the NIH 3T3 Assay / Foci Assay.
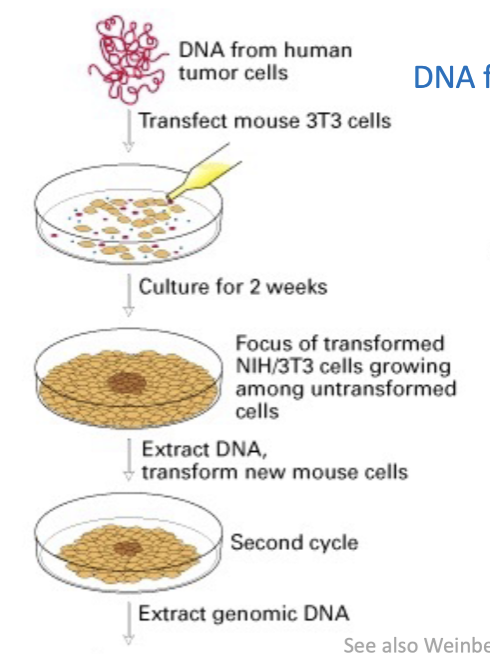
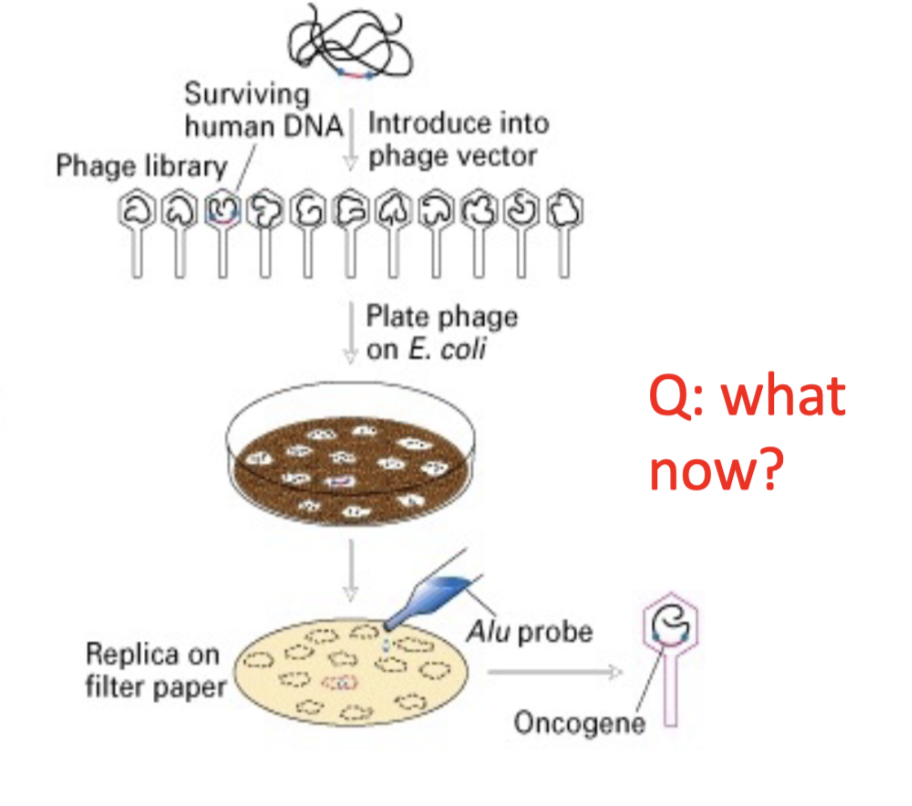
Selection for human sequences using Alu probes
after cloning the oncogene, they searched the data base and matched to H-ras and K-ras
Ras in mitogenic circuitry
So can modulate cell proliferation via gene expression
can also be a part of cell survival
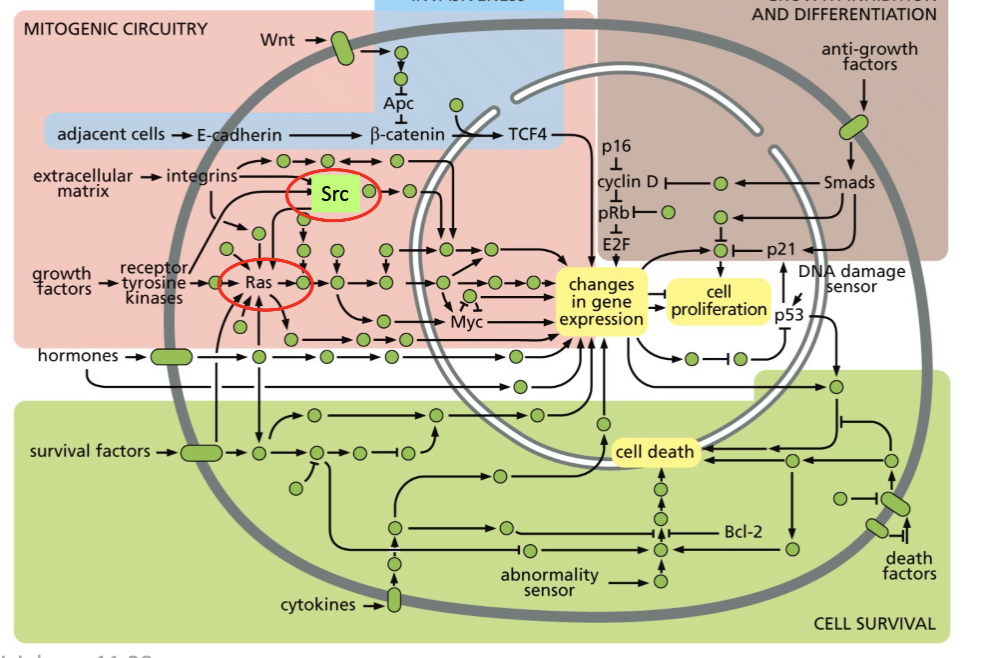
Ras overview
Mutated in approx. 30% of all tumors
50% of colon and 90% of pancreatic cancer
less frequently mutated in other cancers
Why?
Consider cell turnover
Higher proliferation, higher chance of Ras mutations
Microenviornment; inflammation of tissue
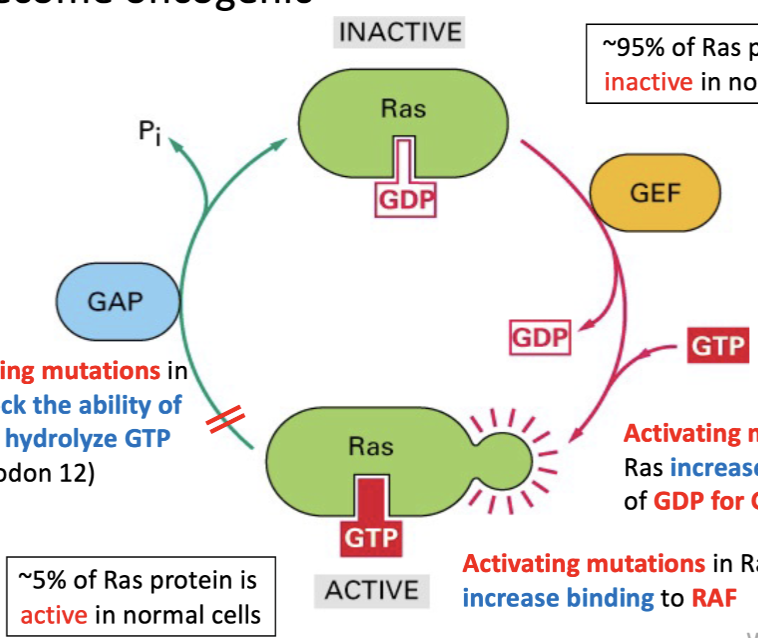
Ras as a Guanine Nucleotide Binding Protein (G-protein):
Binds GDP/GTP.
Cycles between inactive (GDP-bound) and active (GTP-bound) states.
Transitions are aided by Guanine Nucleotide Exchange Factors (GEF) and GTPase Activating Proteins (GAP).
GEF promotes GDP to GTP exchange.
GAP stimulates GTP hydrolysis.
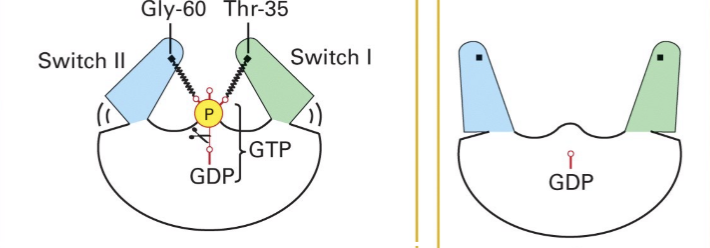
Domains of Ras involved in on/off
Switch I & II
bind to the γ-PO4 of GTP in the active "on" state.
Hydrolysis of the PO4 causes Switch I & II to relax, generating the inactive "off" state, GDP bound off-state
Ras mutations to oncogene
95% of Ras protein is inactive
Activating mutations in Ras:
Increase exchange of GDP for GTP.
Increase binding to RAF.
Block GAP's ability to hydrolyze GTP (e.g., codon 12).
Common mutations that converts the normal Ras gene into an actively transforming oncogene
A single point mutation (G>T) in codon 12 results in a Gly>Val change, converting the normal Ras gene into an actively transforming oncogene.
Codons 13, 59, and 61 are also hotspots for point mutations that activate Ras.
Codons are often found in Guanine Nucleotide Binding Pocket
AA substitutions in the effector loop (switch1) (codons 35, 37, 40) affect interactions with downstream effectors of Ras.
Codons 60 and 61 in Switch II are also frequently mutated in cancer.
Guanine Nucleotide Binding Pocket
Activating mutations in Ras are often found in the guanine nucleotide binding pocket (codon 12).
Mutations in codon 12 (GGC → GTC) inhibit GTP hydrolysis, keeping Ras in the active state
Will not be converted to GDP to turn off
Mutations in codon 12
Have been linked to decrease colon cancer survival
in animals: mutations in codon 12 are induced by chemical carcinogens
B[a]P (Benzo[a]pyrene) ==> lung tumor ==> codon 12 mutated
AFB1 (Aflatoxin B1) ==> liver tumor ==> codons 12 and 61 mutated
Alkylating agent (MNU) ==> breast tumor ==> codon 12 mutated
Diethylnitrosamine ==> liver tumor ==> codon 61 mutated
Conservation within members of the Ras family
highly conserved, except for the C-terminal domain (CTD).
The CaaM,S motif at the very C-terminus is conserved, suggesting its importance for Ras function.
Position of Ras in the plasma members
Anchored to the inner plasma membrane
The C-terminus (at CaaM,S) is prenylated, allowing attachment to the plasma membrane.
Prenylation = addition of a prenyl group (like a fatty anchor).
Made up of isoprene units.
Carried out by farnesyl transferase.
How can farnesyl transferase inhibitors be potential anti-cancer drugs?
If the prenylation of C-terminus (which is required for the attachment of RAS, making it oncogenic) is carried out by farnesyl transferase, inhibitors can be potentially anti-cancer
Used to treat leukemias and other diseases
Isoprene
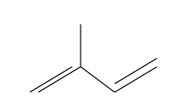
anchors protein into membrane
Farnesyl pyrophosphate (FPP)

Limonene
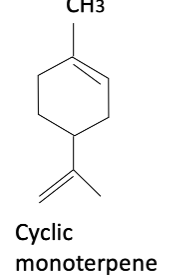
founds in orange and lemon peels, also blocks prenylation
Besides binding GTP and localization of inner plasma members, what else does Ras do?
transduces the signal from membrane-bound GF receptors to a kinase cascade.
SH2 and SH3 domains are involved.
Difference between EGFR and v-ErB
EGFR: Epidermal growth factor →receptor tyrosine
V-ErBb: Viral oncogene
Why is there a difference?
EGFR has a ectodomain that is missing in the v-ErBb
ectodomain is involved in regulation
has a kinase domain
What does EGF binding do do?
Binds to dimeric EGFR causing transphosphorylation
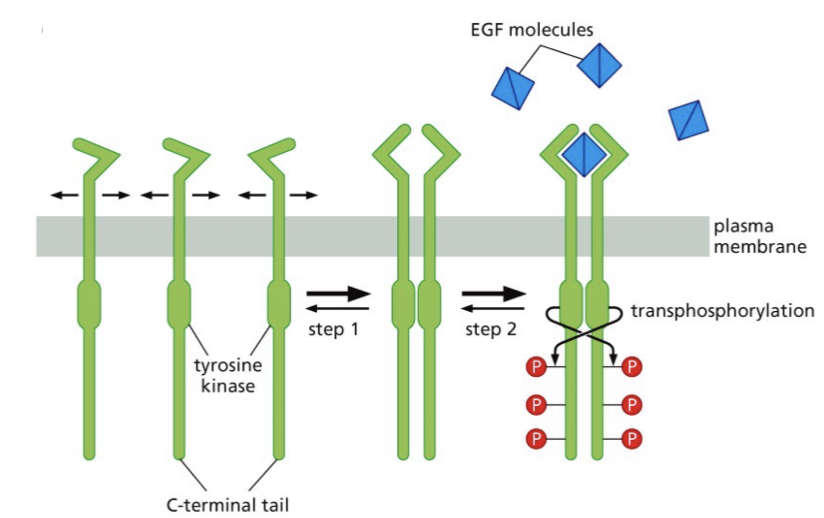
creates active site to phosphorylate proteins like Ras
Ligand-independent firing causing oncogenesis
Mutations can cause the activation of GF receptors without the presence of GF
Absence of ligand-binding domain causing a change in receptor structure allowing for autophosphorylation
Overexpression of receptor, crowing, causing an initiation of phosphorylation
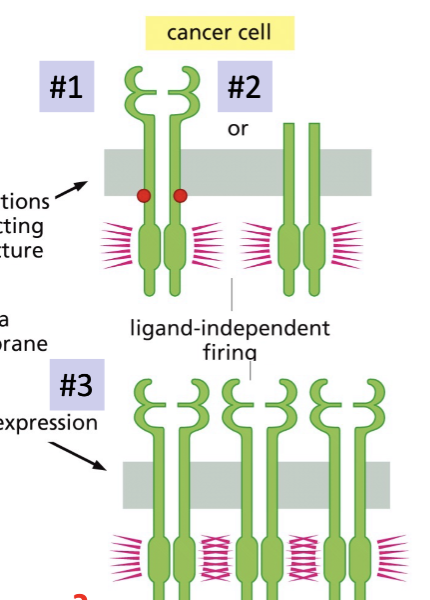
Ligand-independent dimerization
In certain glioblastomas, the ROS RTK gene is fused with the FIG gene which contains a dimerization domain
In the fusion protein, FIG causes the RTK to dimerize (& autophosphorylate) in the absence of GFs.
becomes oncogenic
Two ways that GF receptor can become oncogenic?
Ligand-independent firing
Ligand-independent dimerization
What happens after receptor tyrosine kinases are activated?
GFs activate GF receptors within seconds.
Ras is activated within minutes.
5 minutes after adding EGF to cells, Ras is activated.
GF receptors signal to Ras
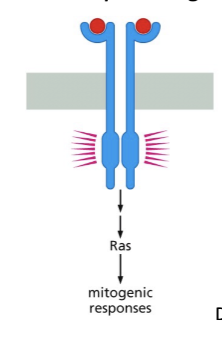
Mitogen
Anything that stimulates a cell to proliferate
MAPK Pathway
mitogen-activated protein kinase. A mitogen stimulates cell proliferation.
Activated Ras is a key initiator of several signaling pathways. The best-characterized is the MAPK pathway.
How is MAPK activated?
Active (GTP-bound) Ras recruits Raf (rapidly accelerated fibrosarcoma to/Ser/Thr kinase) the membrane.
Hydrolysis of Ras-GTP to Ras-GDP releases active Raf then dissociates from Ras
Raf goes on to activate MEK through phosphorylation
MEK is a dual-specificity kinase (Tyr & Ser/thr) that phosphorylates MAP Kinase (MAPK) – a ser/thr kinase.
What occurs after MAPK is activated?
Dimerization of MAP kinase, which allows phosphorylation of p90RSK) in the cytosol, which translocate to the nucleus and phosphorylates SRF and TCF
P-TCF and P-SRF bind the SRE in the promoter of the c-Fos gene (c-Fos is an oncogene).
(SRF = serum response factor; SRE = serum response element. Serum: full of GFs/mitogens)
Basically one kinase activating another
Scaffolding proteins
Hold kinases in place in the ST pathway
promotes specificity and efficiency of the pathway
only the appropriate targets are phosphorylates in an efficient manner
The concentration of both the kinases and the scaffolding proteins needs to be just right for a normal ST pathway.
Free diffusion
No scaffolding proteins
would take time for signal to find its target
cannot explain the specificity and efficiency of ST pathways
Local adaptors
Anchor the scaffolding protein, and hence the kinases, to
the inner plasma membrane (where Ras resides)
SAPK pathway/Stress JNK Pathway
ST pathway activated by stress (UV, ROS, osmolarity changes, heat shock, protein synthesis inhibitors, inflammatory cytokines)
JNK is the hey kinase in this pathway
Can result in
Apoptosis.
Immune activation.
Transformation (c-Jun).
Adaptation to stress.
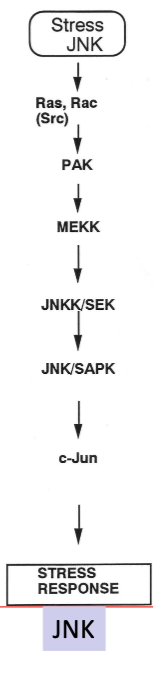
What else can activate MAPK pathway other than GF
Phorbol esters (tumor promoter) can activate protein kinase C (PKC)
ser/thr kinase activated by Ca++ and diacylglycerol (and TPA)
phosphorylates and activates Raf, a member of a family of PKCs; also phosphorylates c-Jun and other TFs. Overexpression can transform cells.
Both ionizing and non ionizing (UV) radiation can stimulate the ST pathway by activating growth factor receptors
TPA: Tetradecanoyl phorbal acetate
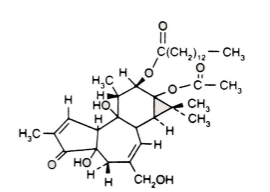
Phosphorylates and activates Raf, a member of a family of PKCs; also phosphorylates c-Jun and other TFs. Overexpression can transform cells.
What are the differences between kinases and oncogenic kinases?
Kinases have regulatory regions that control the activity of the kinase domain
oncogenes kinases lack the regulatory domains