Module 14 - Gene Regulation in Pro/Euk
1/67
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
68 Terms
Gene Regulation
also known as genetic control; determine when, where, and how much a gene is expressed
Every gene is an ___ or ___ region of DNA. Nearby regulatory region(s) are ____.
RNA-coding or transcribed; not transcribed
Transcription factors
DNA-binding proteins that recognize specific sequences within regulatory region(s) near the gene to either activate or repress transcription.
Gene Regulation in Prokaryotes: Operon
an operon is a cluster of structural genes with related functions under the control of a common regulatory system that can respond to changes in environmental conditions.
common in prokaryotes, but rare in eukaryotes
Gene Regulation in Prokaryotes: Operon Function/Mechanism
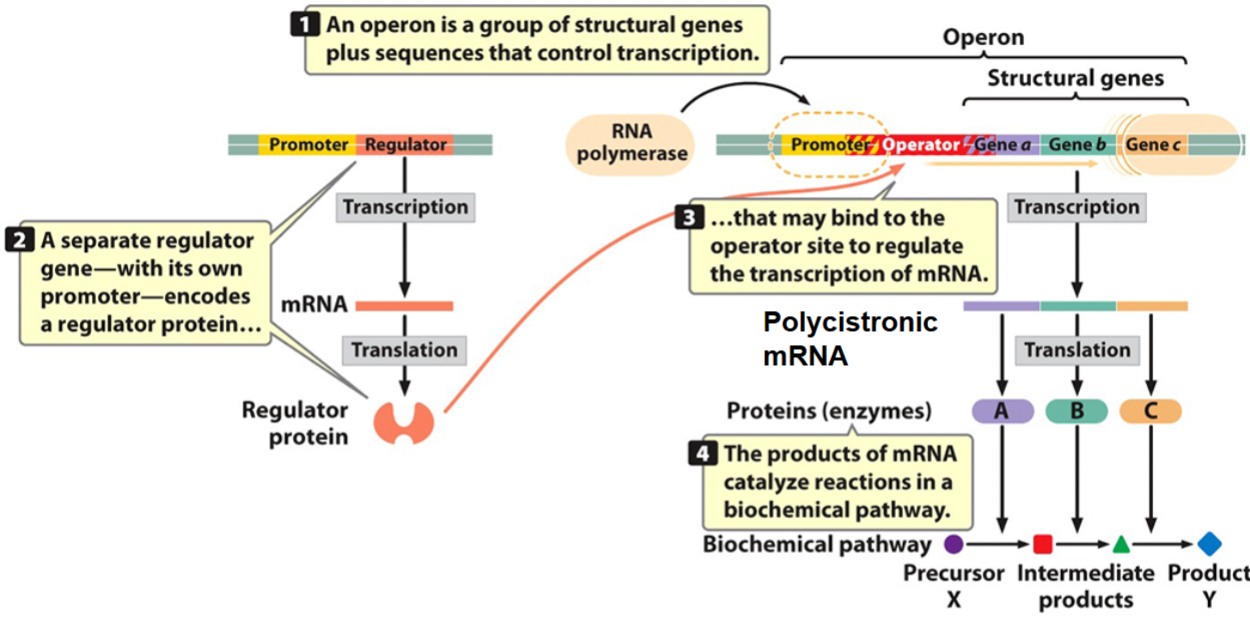
an operon is a group of structural genes plus sequences that control transcription.
a separate regulator gene, with its own promoter, encodes a regulator protein…
…that may bind to the operator site to regulate the transcription of mRNA.
The products of mRNA catalyze reaction in a biochemical pathway.
The lac operon
the products of the structural genes of the lac operon utilize lactose for energy.
genes will only be expressed if lactose is available.
glucose is the preferred energy source for the cell: genes of the lac operon are expressed only if glucose is absent and lactose is available.
Products of the Structural Genes of the lac Operon: b-galatosidase
lacZ gene product
an enzyme that breaks down lactose (a dissacharide) into galactose and glucose.
can also isomerize lactose into allolactose
Products of the Structural Genes of the lac Operon: permease
lacY gene product
a membrane transporter of lactose
facilitates the entry if lactose into the bacterial cell
Products of the Structural Genes of the lac Operon: transacetylase
lacA gene product
not involved in lactose metabolism
involved in the removal of by-products of lactose digestion from the cell.
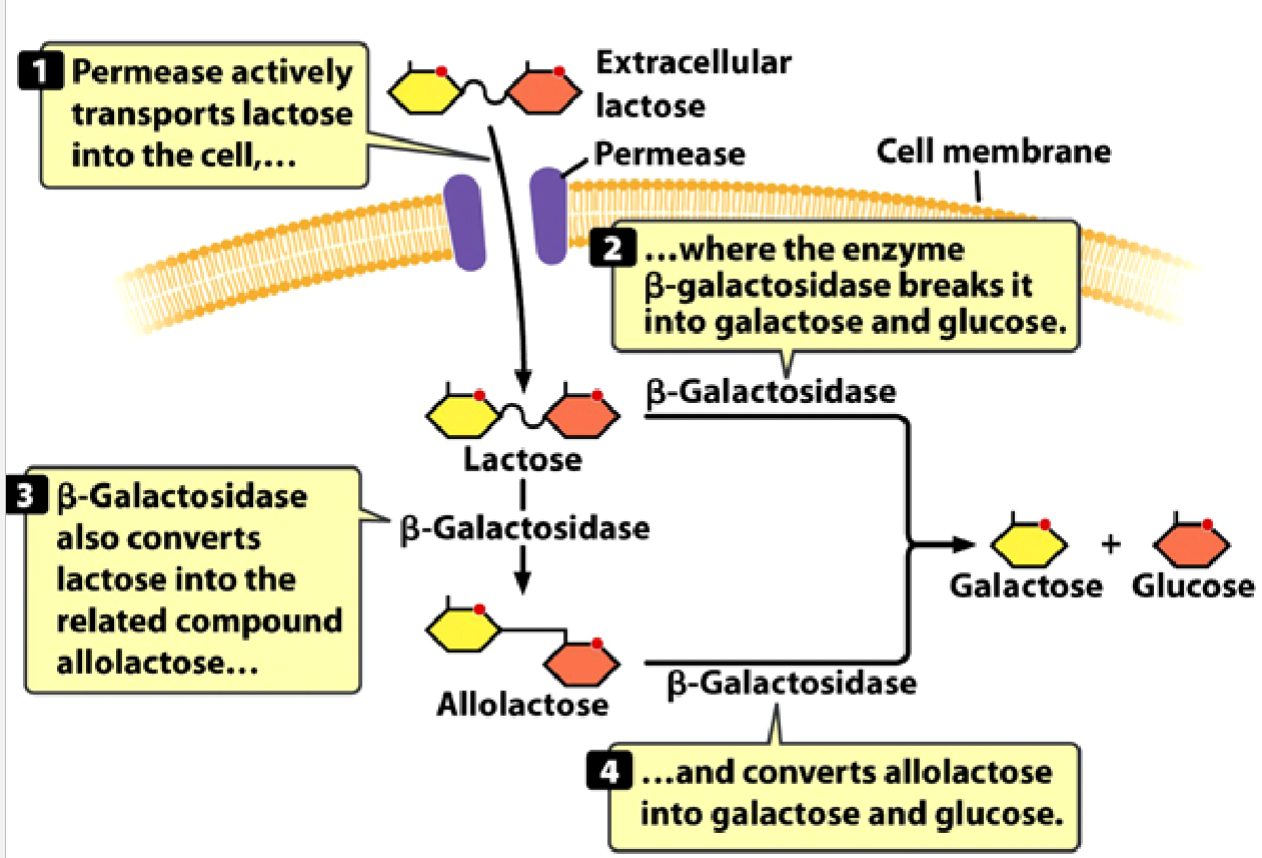
Functions of b-galactosidase and permease:
permease actively transports lactose into the cell…
…where the enzyme b-galactosidase breaks it into galactose and glucose
b-galactosidase also converts lactose into the related compound allolactose…
…and converts allolactose into galactose and glucose.
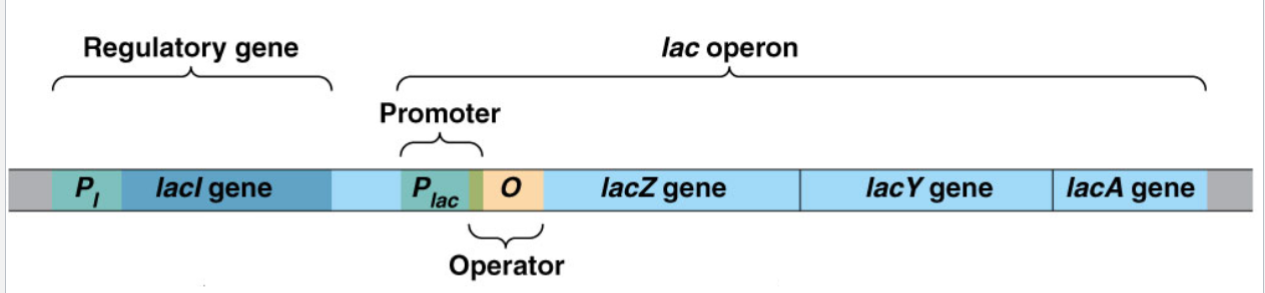
Chromosomal Components of the lac operon:
Structural genes under operon control: lacZ, lacY, and lacA.
Control Regions:
lacP (promoter/Plac): binding site for RNA polymerase
lacO (lac operator): binding site for the repressor protein; overlaps lacP
Regulatory Gene:
lacI (i gene): codes for repressor protein.
PI: promoter for lacI
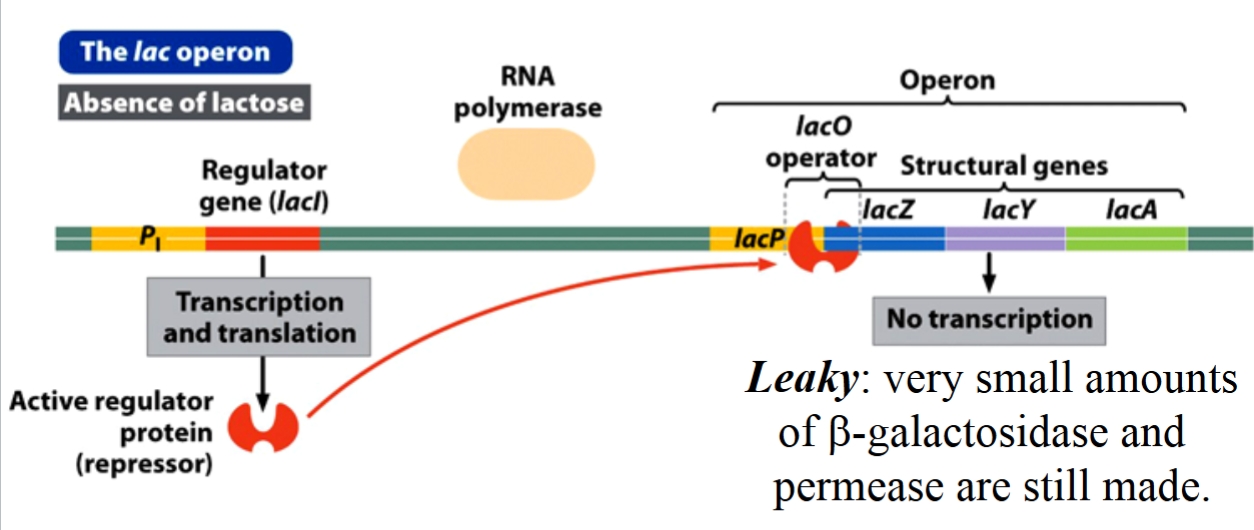
State of the lac Operon in the Absence of Lactose
the regulator protein (a repressor) binds to the operator, blocking RNA polymerase from binding to the promoter.
no expression of the structural genes (repression of gene expression).
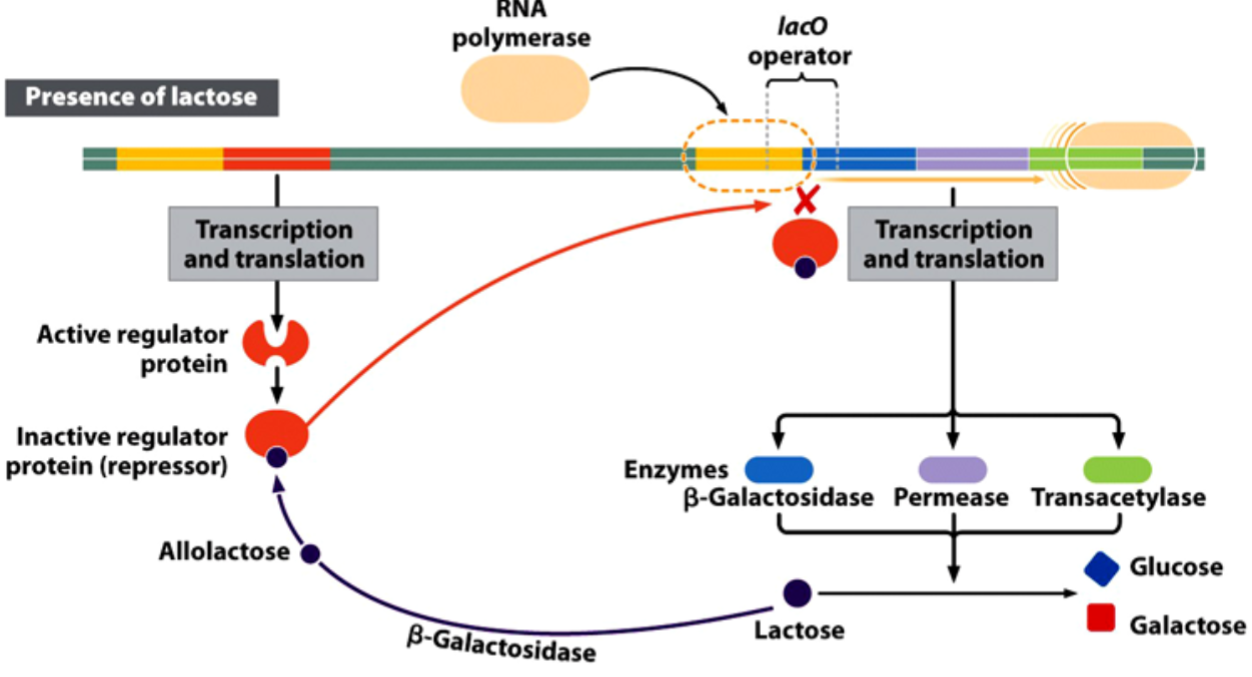
State of the lac Operon in the Presence of Lactose
the operon inducer is allolactose, a lactose metabolite that inactivates the repressor protein.
activates the expression of the genes needed for the use of lactose as an energy source.
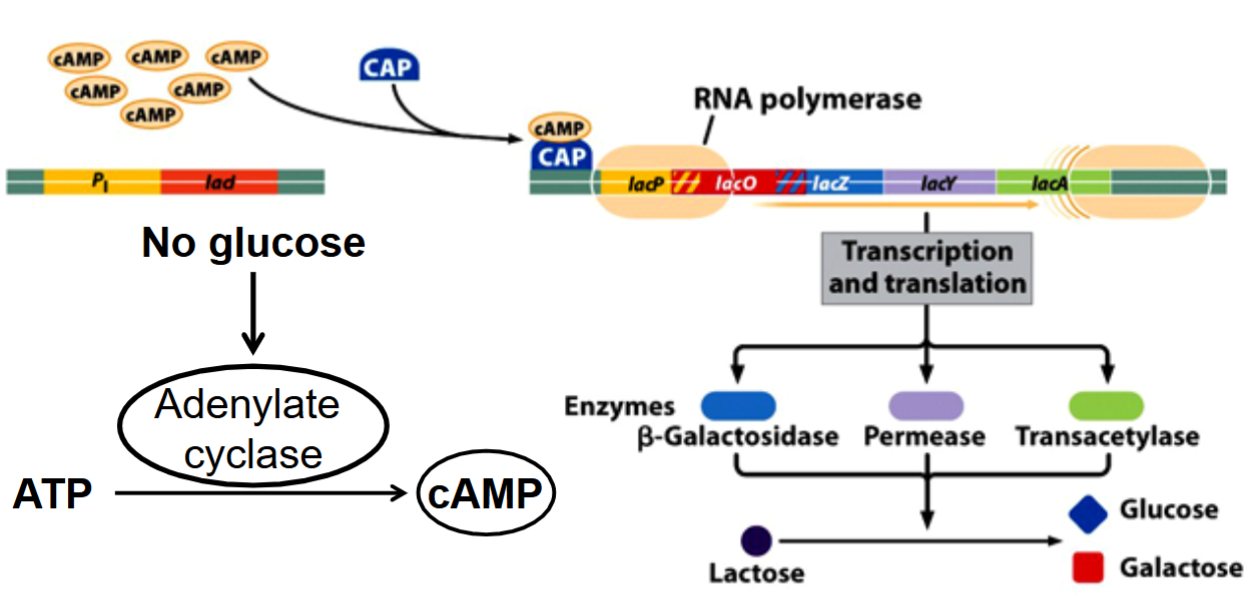
Catabolite Repression of the lac Operon:
If glucose is available, the lac structural genes are off, even if lactose is present; the binding of RNA polymerase to the promoter occurs only if glucose is absent.
Effect of Glucose Levels on Cap Activity:
the activity of the enzyme adenylate cyclase, which catalyzes the hydrolysis of ATP into cAMP + PP, is induced when glucose is absent.
cytoplasmic levels of cAMP increase
cAMP activates the catabolite activated protein (CAP)
the activated CAP binds to a site next to the lac promoter and facilitates the binding of RNA polymerase to it.
cAMP (Cyclic adenosine monophosphate)
the catabolite that binds to catabolite activated protein (CAP) to activate it.
State of the lac Operon in the Absence of Glucose:
catabolite activated protein (CAP) is necessary for the stable binding of RNA polymerase to the lac promoter.
CAP is only active when glucose is absent.
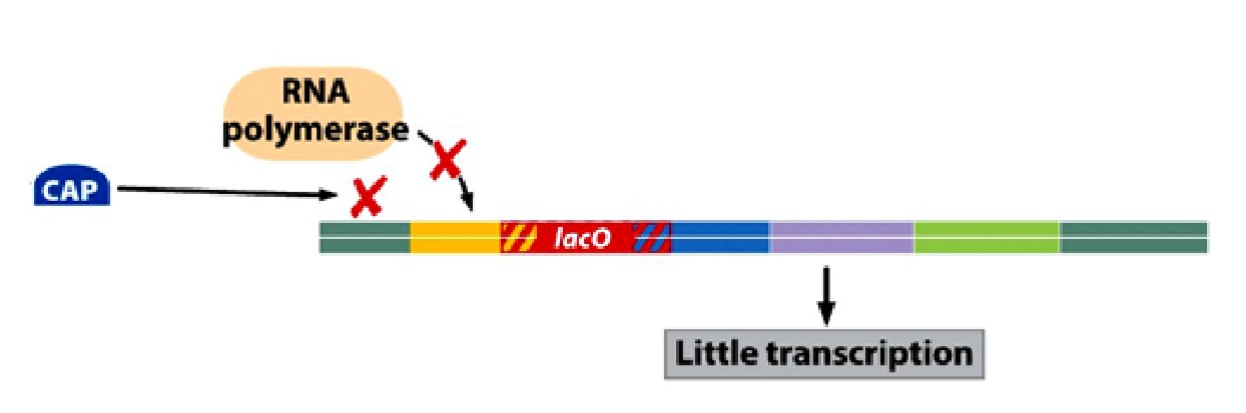
State of the lac Operon in the Presence of Glucose:
cAMP levels decrease, and CAP remains inactive.
RNA polymerase does not bind the promoter efficiently
the operon is OFF
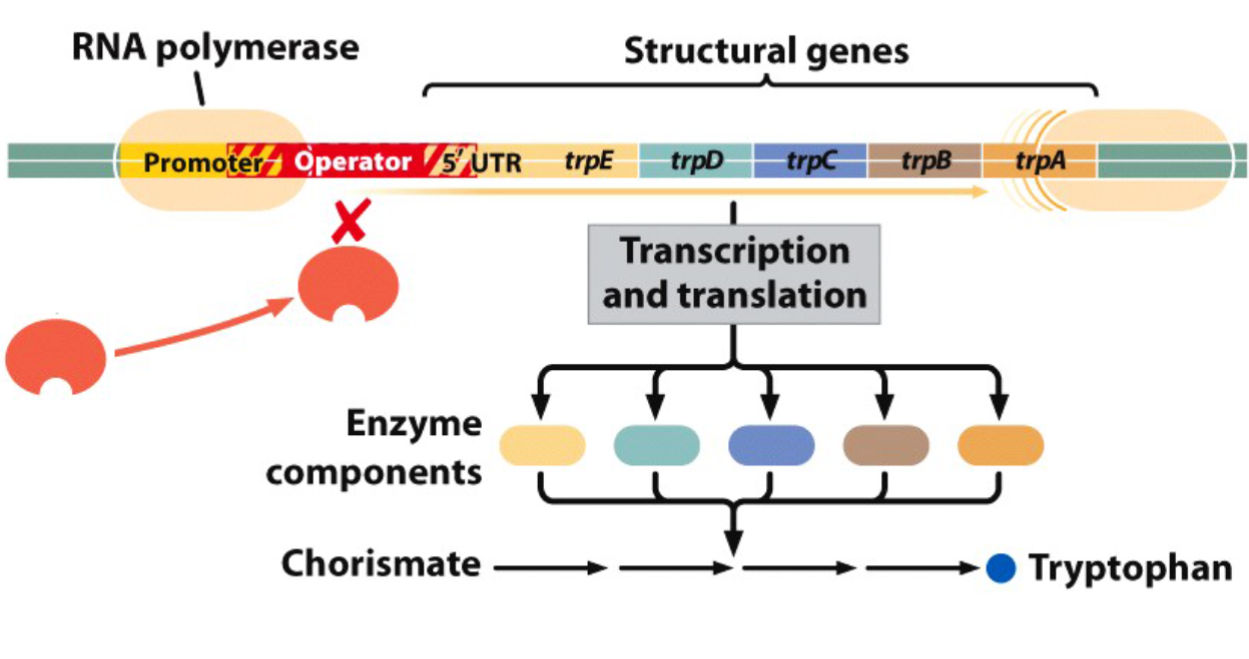
The trp Operon
products of the structural genes of the trp operon are enzymes involved in the biosynthesis of the amino acid tryptophan (Trp).
the structural genes (trpE, trpD, trpC, trpB, trpA) are transcribed as a polycistronic mRNA beginning with the 5’ untranslated region (5’ UTR).
if Trp is absent, the structural genes are expressed.
if Trp is present, the structural genes are turned off by Trp itself

Other Components of the trp Operon
promoter (trpP): binding site for RNA polymerase
operator (trpO): binding site for the repressor protein
repressor gene (trpR): codes for the repressor protein
repressor gene promoter (PR)
The expression of trpR yields an…
inactive repressor protein
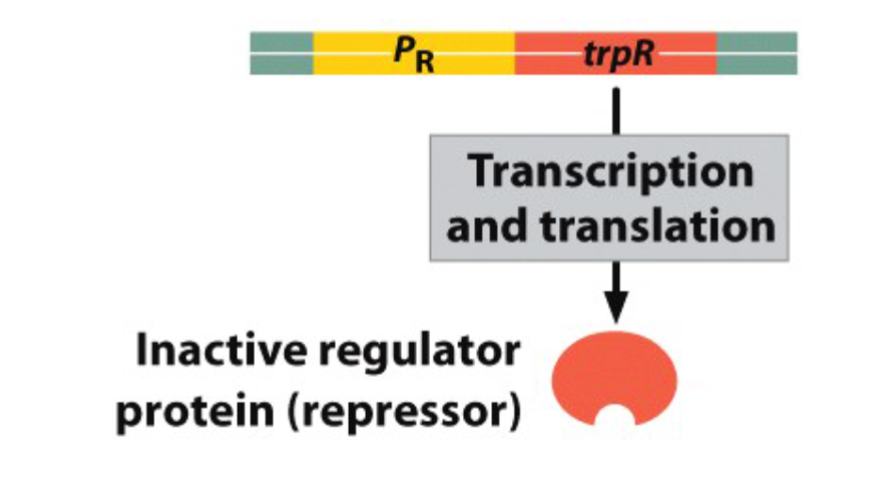
State of trp Operon when Tryptophan is Absent:
the repressor protein remains inactive.
RNA polymerase binds to the promoter and initiates the transcription of the structural genes
Transcription and Translation
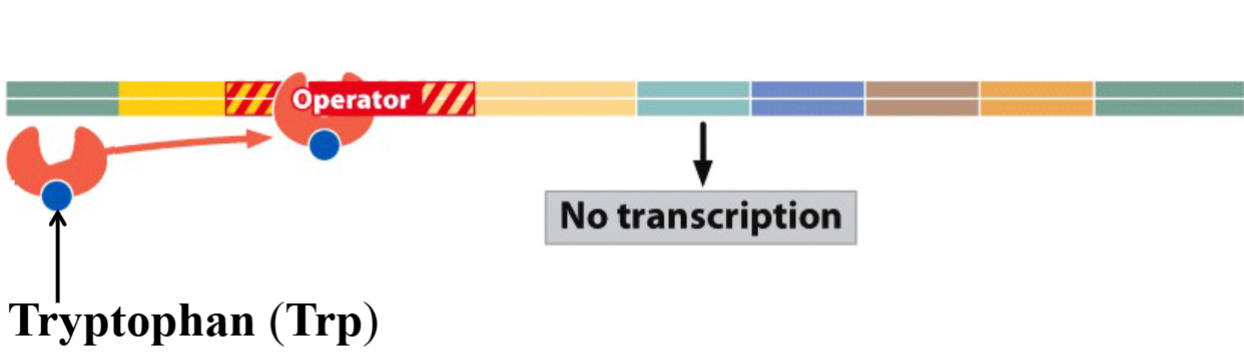
State of trp Operon when Tryptophan is Present:
Trp binds to the repressor protein and activates it.
The binding of the Trp-activated repressor to the operator prevents the binding of the RNA polymerase to the promoter.
No transcription
Electrophoretic Mobility Shift Assay (EMSA)
method of detecting specific DNA-protein interactions
i.e., specific binding of transcription factors and other regulatory regions of the chromosome.
Electrophoretic Mobility Shift Assay (EMSA) involves how many experiments?
5
DNA alone
DNA + cell extract (no Trp)
DNA + cell extract + Trp
DNA + cell extract + Trp + anti-lac repressor antibody
DNA + cell extract + Trp + anti-trp repressor antibody
Electrophoretic Mobility Shift Assay (EMSA) involves what test?
low voltage electrophoresis of sample of the contents of each tube.
Electrophoretic mobility shifts occur due to the different sizes of the complexes that form.
RNA Polymerase I
the larger rRNA genes (5.8s, 18s, and 28s)
RNA Polymerase II
all the protein-coding genes
RNA Polymerase III
the small rRNA (5s) genes and all the tRNA genes
Chromatin Remodeling and the Regulation of Transcription:
heterochromatin is transcriptionally inert due to the tight association of DNA with histones, which prevents RNA polymerase from binding to gene promoters.
the ability of the cell to alter the association of DNA with histones is essential to allow gene regulatory proteins and RNA polymerases to bind to DNA.
chromatin remodeling is necessary for exposing regulatory sites, such as promoters, to make them available for transcription factors and RNA polymerase.
Chromatin Remodeling: Mechanism
the chromatin remodeling complex binds to DNA…
… and repositions nucleosomes, exposing a transcription factor binding site.
transcription factors and RNA polymerase bind to DNA and initiate transcription
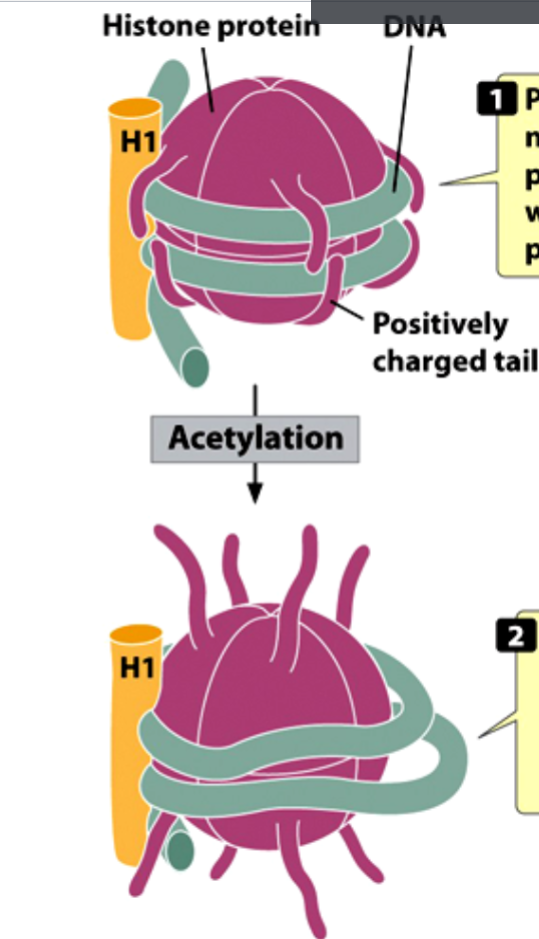
Consequence of Histone Acetylation:
histone acetylation weakens the interaction between basic histones and the acid DNA molecule, causing chromatin recondensation.
HATs and HDACs do not bind to DNA but regulate histone acetylation.
Consequence of Histone Acetylation: Mechanism
positively charged tails of nucleosomal histone proteins probably interact with the negatively charged phosphate groups of DNA.
acetylation of the tails weakens their interaction with DNA and may permit some transcription factors to bind to DNA.
Covalent Modification of Histone:
transcription activators recruit histone acetyl transferases (HATs), causing chromatin recondensation.
transcription repressors recruit histone deacetylases (HDACs), causing chromatin condensation.
HATs act as ___, but they do not bind to ___.
coactivators; DNA
HDACs act as ___, but they do not bind to ___.
corepressors; DNA
Flowering Locus C (FLC) codes for…
a transcription factor that represses flowering but is only expressed if the histones on the locus are acetylated.
Transcription factor refers to…
any transcription regulator protein that binds to a specific DNA sequence
cis-acting elements
DNA sequences that are necessary for the control of transcription.
the regulatory regions of the chromosome: promotors, enhancers, and silencers.
trans-acting factors are…
proteins that are necessary for the control of transcription
transcription factors that bind to the cis-acting elements
Cis-acting Elements: Promoters
the binding sites for the RNA polymerase II transcription initiation complex
always upstream from the transcription initiation site (very close).
recognized by the RNA polymerase II transcription initiation complex (or basal transcription apparatus).
promoters of eukaryotic genes transcribed by RNA polymerase II contain conserved sequence elements such as the TATA box (binding site for the TATA-binding protein/TBP).
Cis-acting Elements: Enhancers
the binding sites for transcriptional activator proteins
required for stimulated transcription
DNA sequences vary widely and are recognized by a large variety of transcription activators.
activation of a gene in any particular cell will depend on whether the cell has the right activators to bind to the gene enhancers.
by facilitating up-regulation, enhancers determine where, when, and how much transcription occurs.
Cis-acting Elements: Silencers
the binding sites for transcriptional repressor proteins
Role of Regulatory Regions: Promoter Alone
A) Promoter Alone: only the basal transcription factors may bind to DNA, and only the basal transcription apparatus may form.
only very low or undetectable transcription can occur
Role of Regulatory Regions: With Right Enhancers
B) transcriptional activators bind to enhancers
stimulated transcription (biologically significant).
Role of Regulatory Regions: C-E
C-E) enhancers can be distant upstream, downstream, or in an inverted position (always on the same chromosome), and they can still be effective is stimulating transcription.
Cannot move the promoter
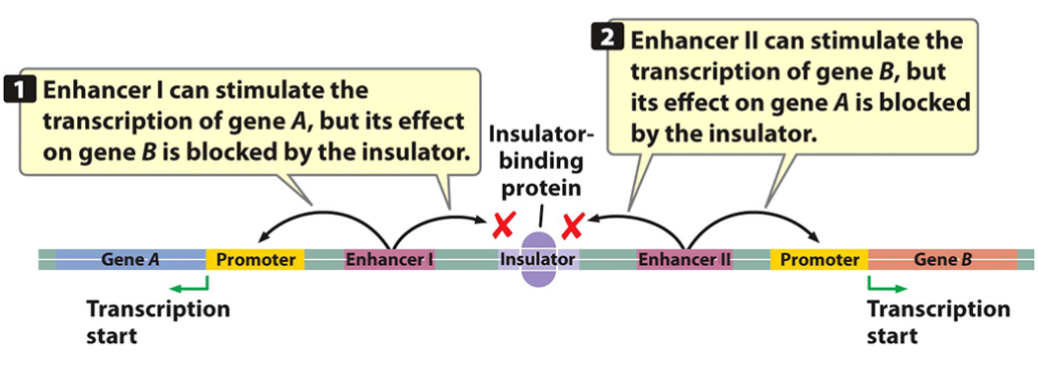
Insulators
enhancer boundary cis elements; DNA sequences that block the effect of enhancers in a position-dependent manner when insulator-binding proteins are bound to them.
prevents enhancers to stimulate the transcription of the wrong genes on the same chromosome.
Purpose of the RNA Polymerase II Transcription Initiation Complex
to direct RNA polymerase II to the correct place on the promoter behind the gene transcription initiation site (+1) because polymerase itself does not recognize any specific DNA sequences.
TFIIs
transcription factors of polymerase II
TFIID complex
transcription factor D of RNA polymerase II
TBP
TATA-binding protein, part of the TFIID complex
Transcriptional activators are necessary for…
stimulated, or biologically significant, levels of transcription.
Transcriptional activators bind to the…
upstream and, sometimes, downstream sequence elements (the enhancers)
RNA Polymerase II Transcription Initiation Complex (Basal Transcription Apparatus) contains:
TFIID
other TFIIs
RNA Polymerase II
DNA loops to allow transcriptional activators, bound to enhancers, to interact with the complex.
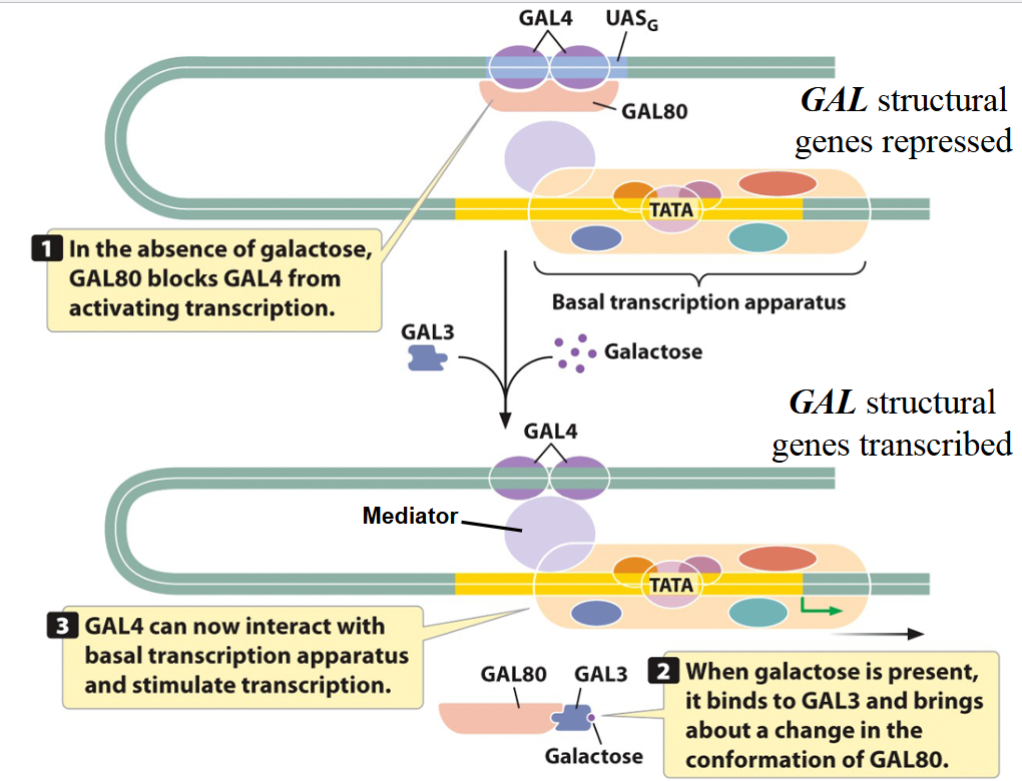
Induction of the GAL Structural Genes
the products of the GAL structural genes in yeast are
proteins that metabolize the sugar galactosethe transcription of these genes requires the enhancer UASG
(upstream activating sequence of GAL genes) which is
permanently occupied by the transcription activator GAL4in the absence of galactose, the GAL80 protein binds to
GAL4 and prevents it from activating the transcription of
the GAL structural geneswhen galactose is available, it activates the GAL3 protein,
which interacts with GAL80 to displace it and expose the
GAL4 trans-activating domaina mediator protein that interacts with both GAL4 and the
transcription initiation complex is required
GAL Structural Genes: Mechanism
in the absence of galactose, GAL80 blocks GAL4 from activating transcription.
when galactose is present, it binds to GAL3 and brings about a change in the confirmation of GAL80.
GAL4 can now interact with basal transcription apparatus and stimulate transcription.
Barbara McClintock pioneered the field of…
cytogenetics
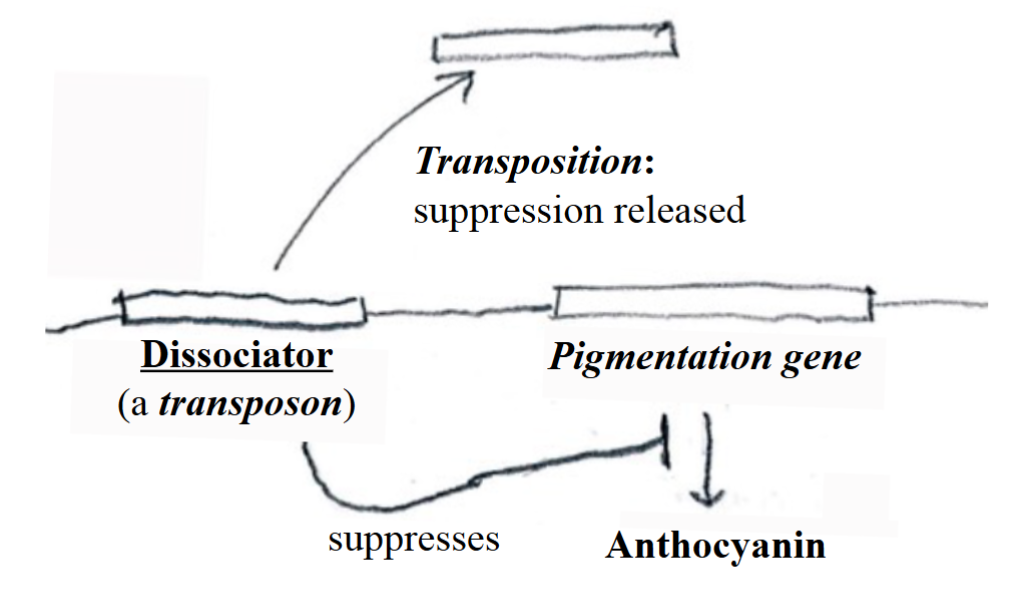
Negative Regulation by Transposons (Barbara McClintock’s transposition model)
the expression of a gene that is necessary for the synthesis of the pigment anthocyanin can be suppressed by a silencer that she named Dissociator. It is a transposon: a segment of DNA that can move from one location on a chromosome to another.
If Dissociator is transpositioned, the suppression of the
pigmentation gene is released, and anthocyanin can be made.
transposon
a segment of DNA that can move from one location on a chromosome to another
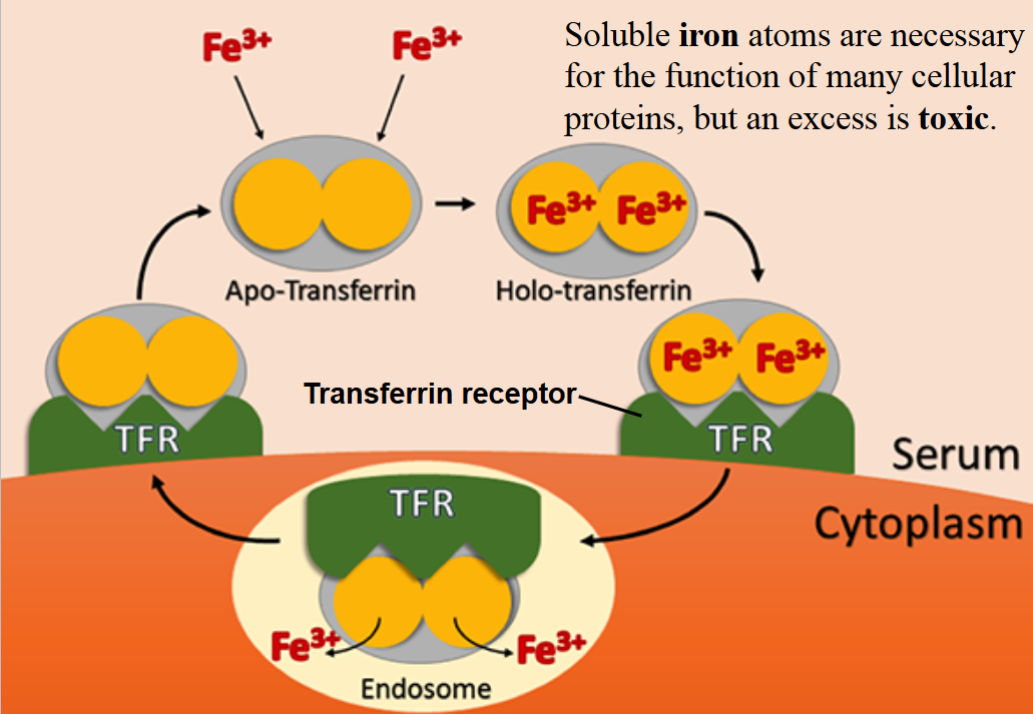
Transferrin System of Iron Transport
iron ions (Fe3+) are transported in the bloodstream by the protein Transferrin as the Transferrin/iron complex.
the Transferrin receptor (TFR), present on all cells, interacts with the holo-transferrin and transports it into the cell, where iron is then released.
•if levels of free cytoplasmic iron increase too much, the cell synthesizes Ferritin, a protein that sequesters iron atom.
Translational Regulation of the Transferrin Receptor: Absence of Free Iron
the iron regulatory protein binds to the iron response element (IRE) at the 3’ UTR of the Transferrin receptor mRNA, forming a stem loop structure that stabilizes the mRNA to promote translation.
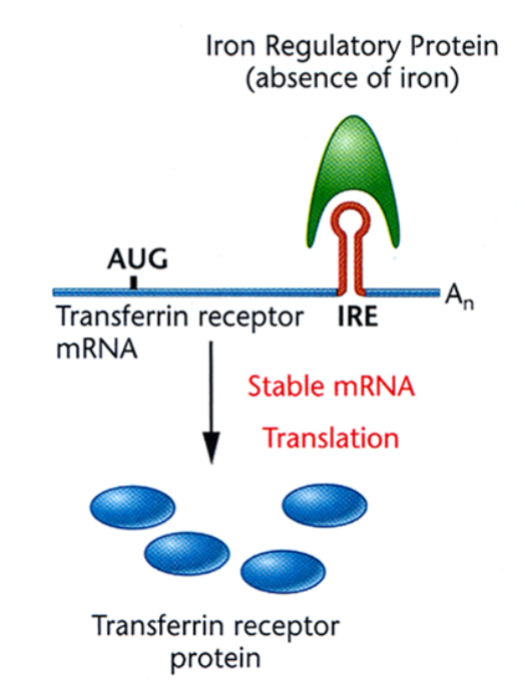
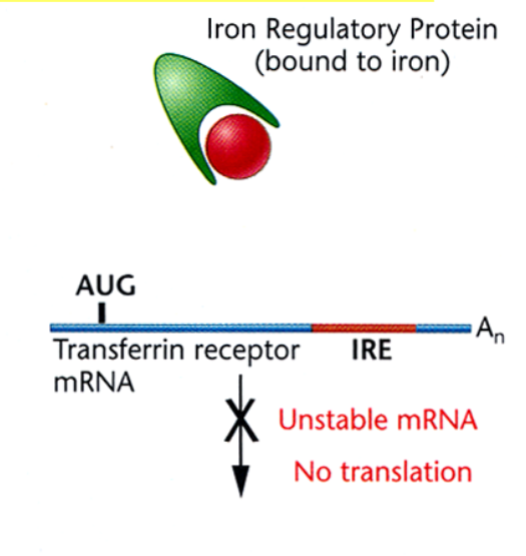
Translational Regulation of the Transferrin Receptor: Presence of Free Iron
the iron atoms bind to the iron regulatory protein and prevents it from binding to the IRE, causing the destabilization of the Transferrin receptor mRNA and disrupting protein synthesis (prevent excess of transport of iron).
Translational Regulation of Ferritin: Absence of Free Iron
the iron regulatory protein binds to the iron response element (IRE) at the 5’ UTR of the Ferritin mRNA, forming a stem-loop structure that, in this case, inhibits translation.
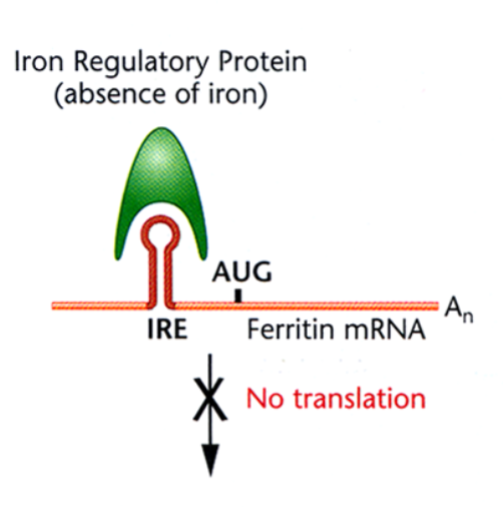
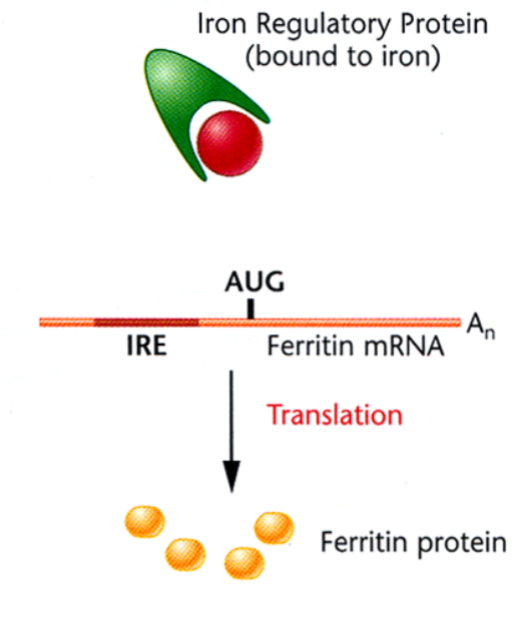
Translational Regulation of Ferritin: Presence of Free Iron
the iron atoms bind to the iron regulatory protein to prevent the formation of the stem-loop structure to allow Ferritin synthesis.
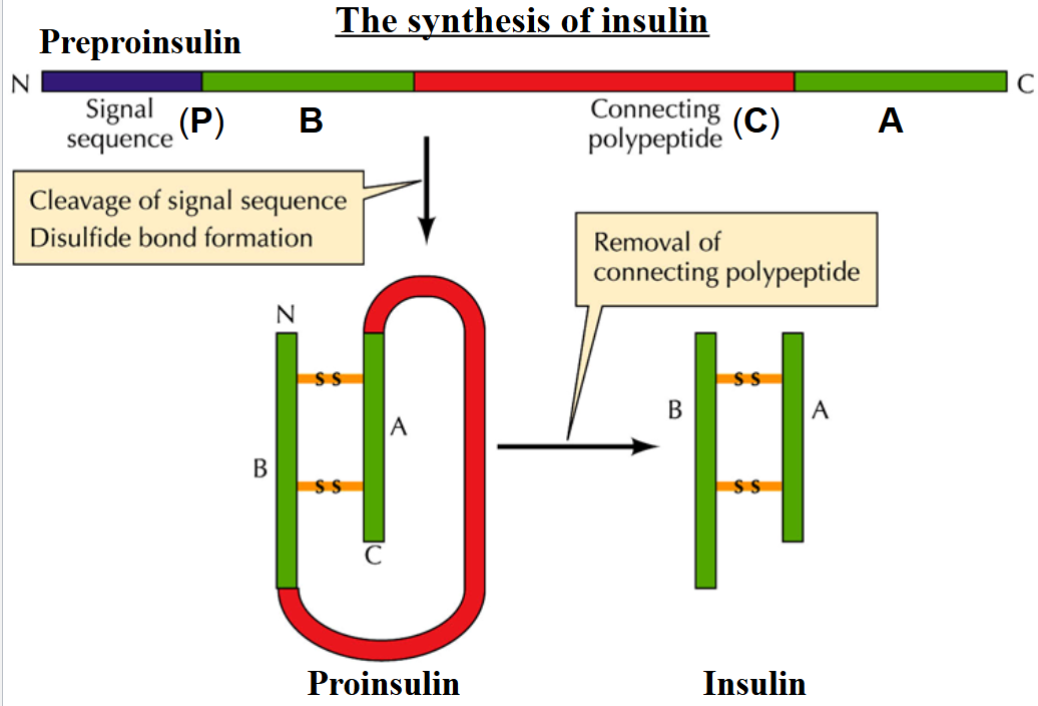
Post Translational Regulation: Proteolytic Cleavages
example: synthesis of insulin
cleavage of signal sequence + disulfide bond formation
removal of connecting polypeptide
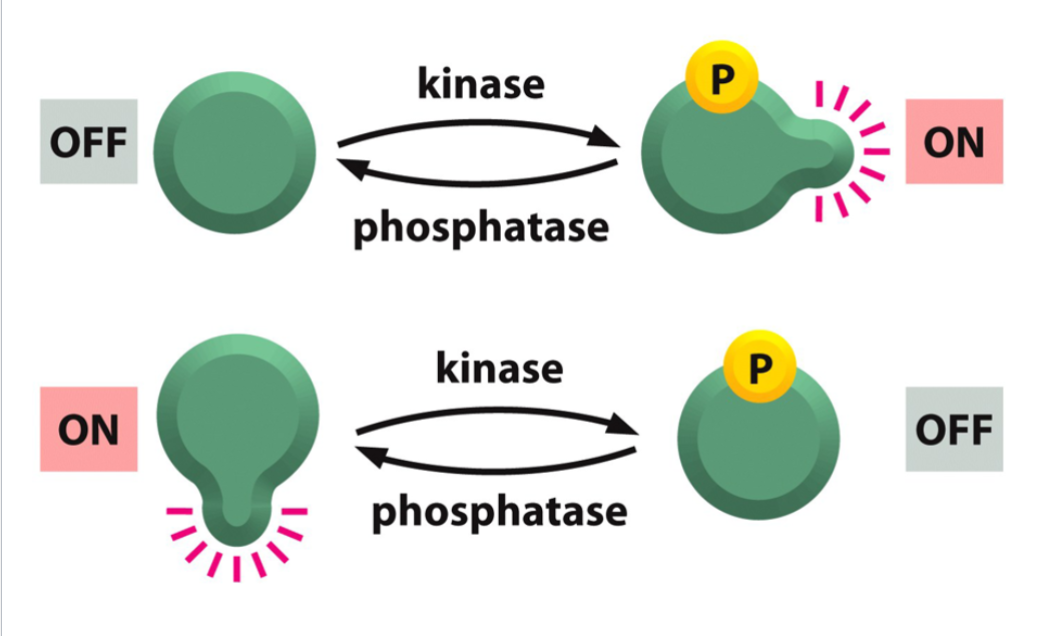
Post Translational Regulation: Protein Phosphorylation
a covalent modification necessary for the activation or deactivation of many proteins
Protein Kinases
enzymes that transfer phosphate groups from ATP to proteins; comprise a very large family of enzymes.
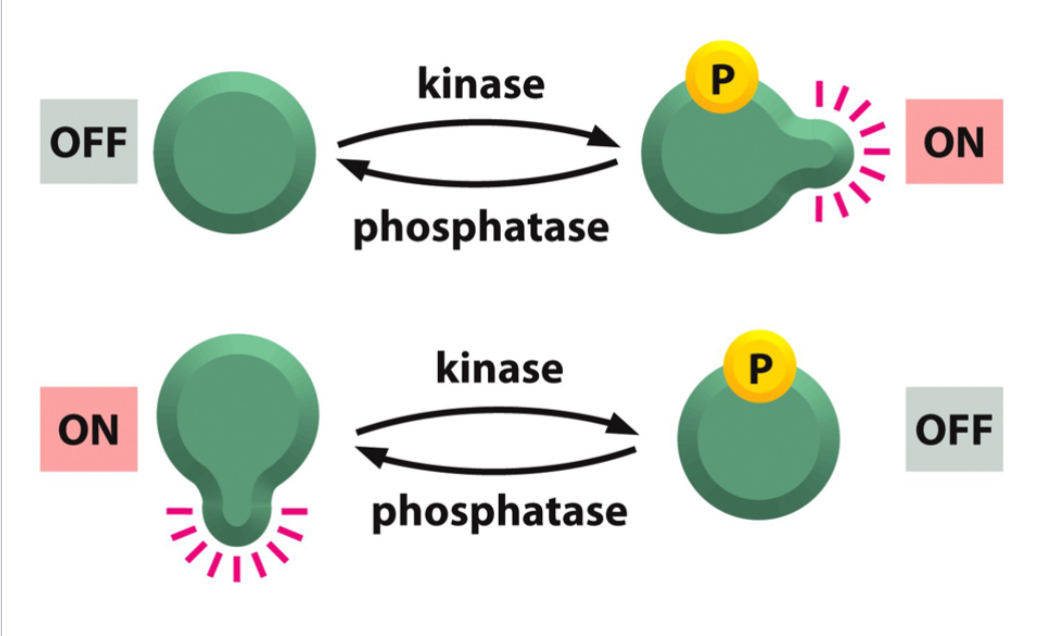
Protein Phosphates
enzymes that remove phosphate groups from phosphorylated proteins.