Nucleic acids
1/28
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
29 Terms
What is the structure generally of nucleotides and what is nucleic acid
Nucleic acids are polymers of monomers of nucleotides.
A molecule containing many nucleotides = polynucleotides.
A nucleotide has 3 components which are combined in condensation reaction:
A phosphate group which is the same structure in all nucleotides
A pentose sugar - eg. Ribose in RNA and deoxyribose in DNA
An organic base or 'nitrogenous base'.
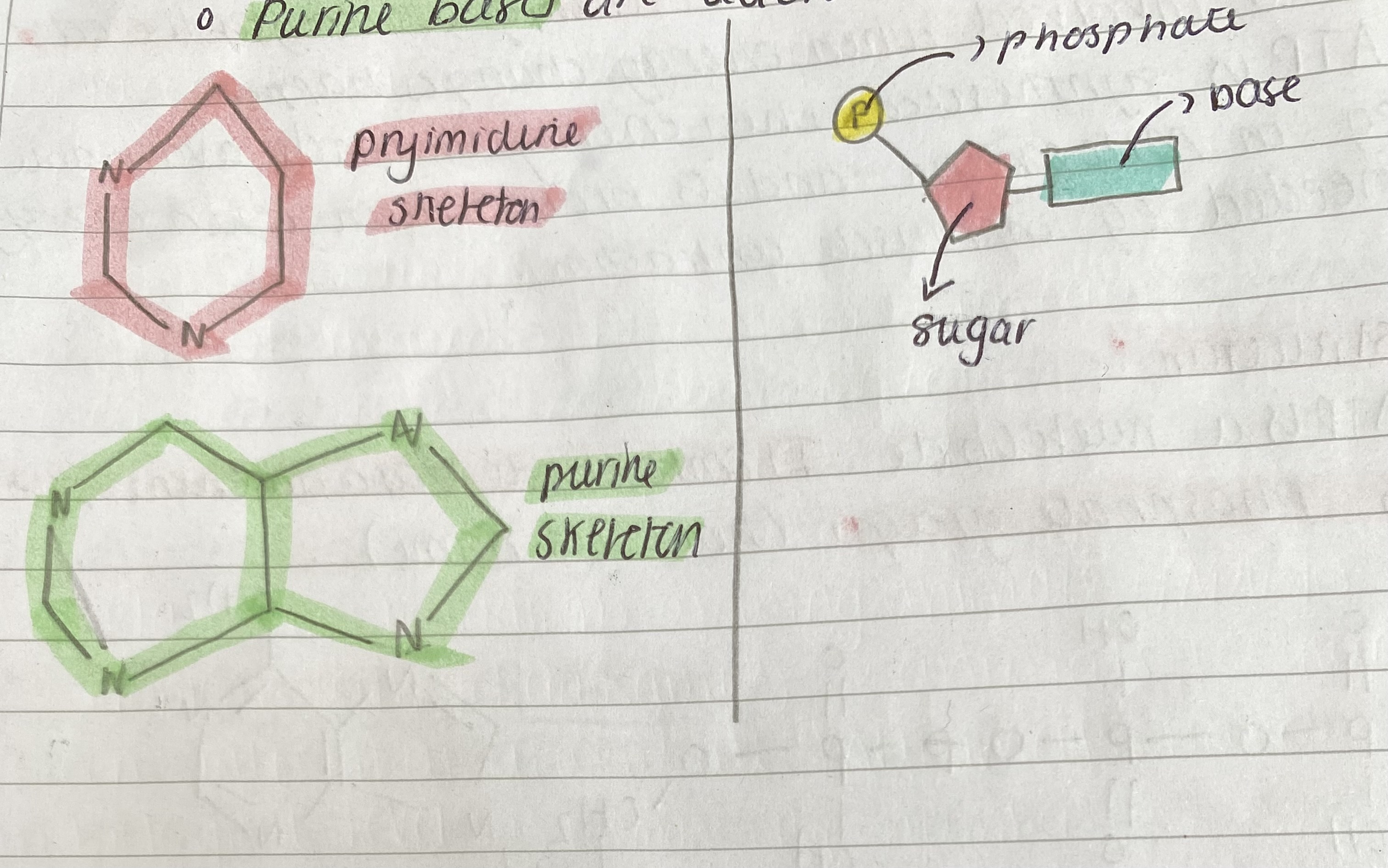
There are two groups of organic bases
o Pyrimidine bases are Thymine, cytosine and Uracil (U).
o Purine bases are adenine and guanine
Intro to ATP - chemical energy and biological processes
In biological systems it is chemical energy that makes the changes because chemical bonds must make or break for reactions to occur.
Heterotropic organisms eg animals derive from chemical energy from food whereas autotrophic organisms like green plants derive it from light energy, which they convert into chemical energy via photosynthesis.
What is ATP And how is it used as an ‘energy currency’
ATP as an energy carrier
Organisms store chemical energy mainly in lipids and carbohydrates but adenosine triphosphate (ATP) makes their energy available when it is needed.
We need to make and break down about 50kg of ATP per day but the body only contains 5g of ATP so is not an energy store.
It is sometimes called the energy currency of the cell as it is involved when energy changes happen. The idea of energy currency comes from the idea that it is involved in all exchanges of all living organisms. It is synthesised when energy is made available So it’s exchanged. Transfers energy from place of release to others. It consists of 3 phosphates, sugar ribose and the organic Adosine base. Easily reversible condensation + hydrolysis
ATP is synthesized when energy is made available eg in mitochondria and is broken down when energy is needed eg in muscle contraction.
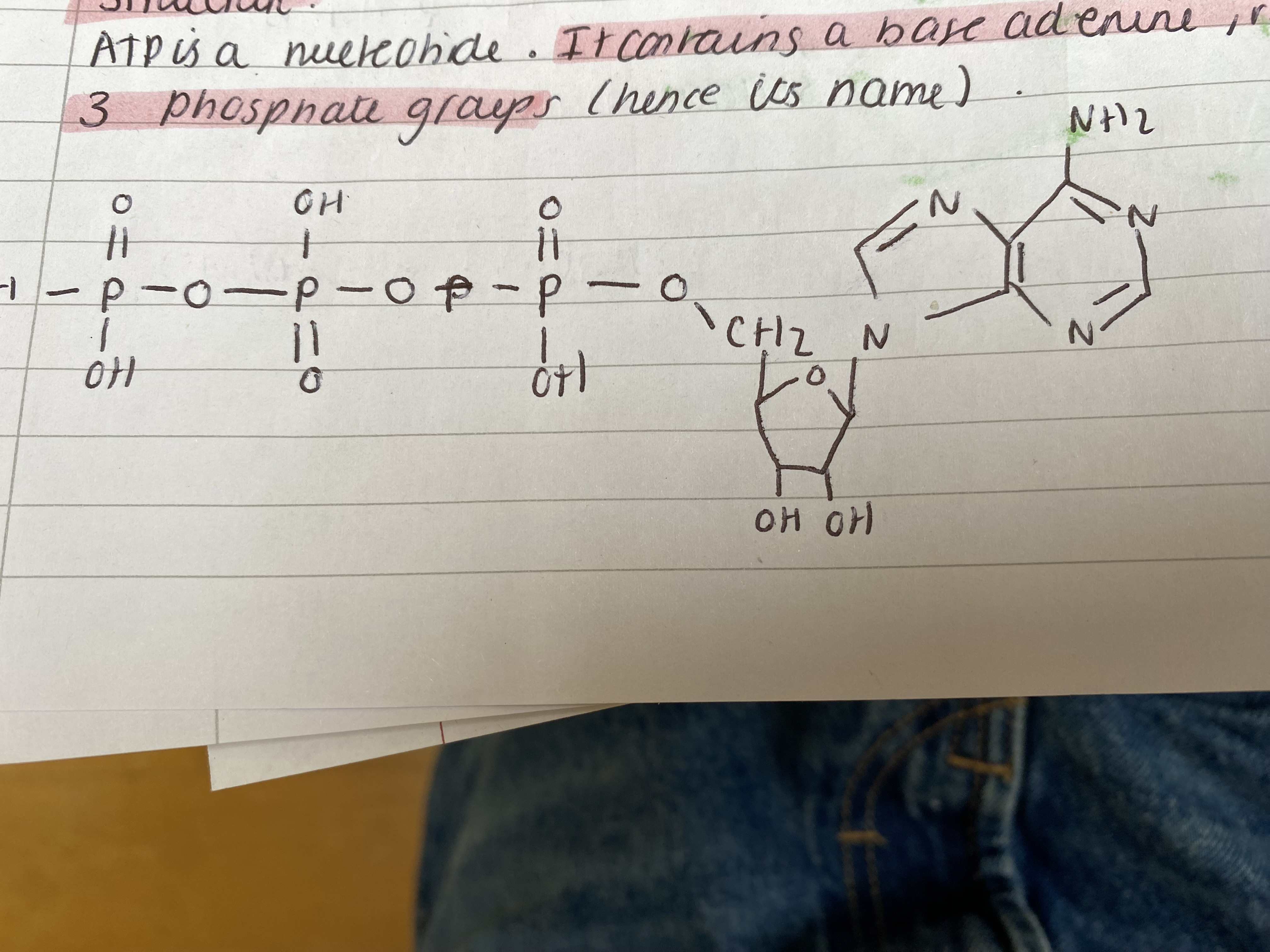
Explain ATP structure
structure
ATP is a nucleotide. It contains a base adenine, ribose and 3 phosphate groups (hence its name).
Explain how ATP releases energy
ATP and energy
When energy is needed in living organisms ATPase (enzyme) hydrolyses the bond between the 2nd and 3rd phosphate groups in ATP and removing the third phosphate leaving only 2. The ATP to this forms adenosine diphosphate (ADP) and an inorganic phosphate ion with the release of chemical energy it catalysed by ATP hydrolase Every mole of ATP hydrolysed releases 30.6 kJ when the bond is broken. A reaction that releases energy like ATP hydrolysis is exogenic.
ATP + water → (reversible) ADP + Pi triangle H = 30.6KJ mol-1
The condensation reaction requires energy (input endergonic) and energy not synthesised needs 30.6 kJ.
The addition of phosphate to ADP is called phosphorylation.
ATP transfers free energy from energy-rich compounds like glucose to cellular reactions where it's needed. But energy transfers are inefficient and some energy is always lost as heat. The uncontrolled release of energy from glucose would produce a temperature increase that would destroy cells. Instead living organisms release energy gradually in a series of small steps called respiration to produce ATP.
Explain ATP advantages as a supplier of energy
ATP as a supplier of energy
There are several advantages to having ATP as an intermediate in providing energy compared with glucose directly.
• The hydrolysis of ATP into ADP involves a single reaction that releases energy immediately. The breakdown of glucose requires many intermediates and it takes longer for energy to be released.
only one enzyme is needed to release energy from ATP unlike glucose.
ATP releases in small amounts when and where it is needed glucose releases large amounts of energy at once.
ATP provides a common source of energy for many different chemical reactions, increasing efficiency and control by the cell.
Explain the roles of ATP
The roles of ATP:
ATP provides the necessary energy for cellular activity.
metabolic processes - to build large complex molecules eg. DNA synthesis from smaller simpler mols eg nucleotides
active transport - to change the shape of carrier proteins in membranes and allow ions to move against conc. g
movement for muscle contraction
nerve transmission - sodium potassium ions pumps actively transported sodium and potassium ions across axon membrane
secretion - vesicles - transport & package of secretory products
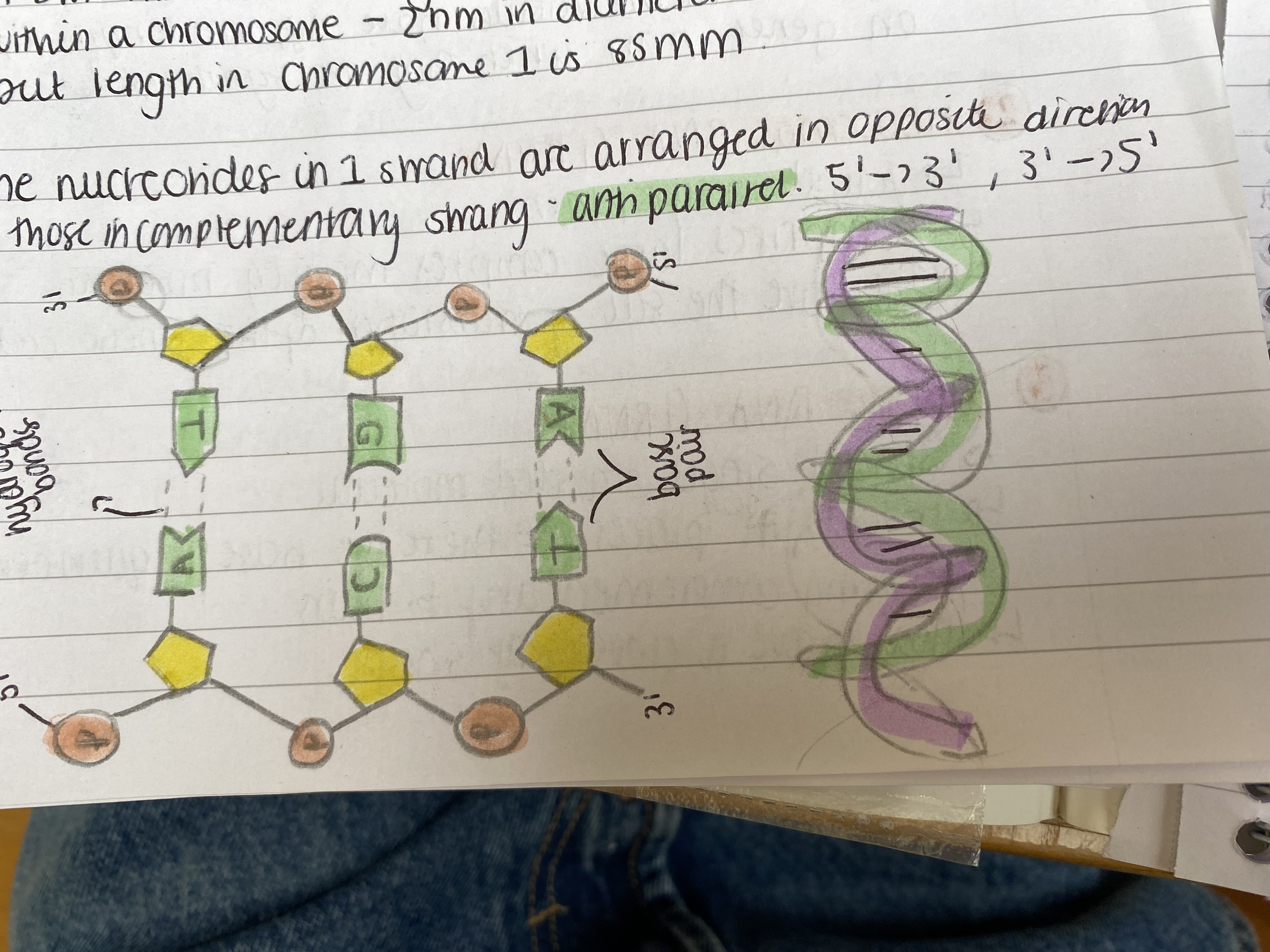
Explain the structure of DNA
The structure of DNA:
DNA has 2 polynucleotide strands wound around each other in a double helix
The pentose sugar in the nucleotide is deoxyribose
There are 4 organic bases: 2 purine (adenine, guanine) and 2 pyrimidines (cytosine and thymine)
phosphate
The deoxyribose and phosphate groups are on the outside or form the 'backbone' - covalent phosphodiester bonds
The bases of the 2 strands face each other and point inwards.
Adenine opposite thymine always and Guanine always opposite cytosine
Hydrogen bonds join bases to 'complementary pairs'The hydrogen bonds are weak as they need to be broken after
These bonds maintain the helix shape
A DNA molecule is long, thin and tightly coiled within a chromosome - 2nm in diameter but length in chromosome 1 is 85mm
The nucleotides in 1 strand are arranged in opposite direction to those in complementary strand - anti-parallel 5' - 3', 3' - 5'
Explain the role of DNA in terms of its structure
The structure of DNA means its suited to its functions
• A very stable molecule and its information is generally essentially unchanged from generation → generation
• It is a very large molecule and therefore carries a large amount of information
• As the base pairs are on the inside of the double helix inside backbone → genetic info is protected.
• The two strands are able to separate due to Hydrogen bonds.
How was DNA structure discovered - history
DNA:
Watson and Crick proposed the molecular structure of DNA in 1953. They used the information obtained by many scientists including Franklin and Wilkins to build the 3D DNA model.
Franklin obtained the photo of DNA by putting a beam of x-rays scattered by a crystal to make a pattern on a film which depends on the crystal structure. The photo indicates the distance between atoms + DNA structure.
Explain the structure of RNA
The STRUCTURE of RNA
• It is a single-stranded polynucleotide
• RNA contains the pentose sugar ribose
• RNA contains the purine bases adenine, guanine and the pyrimidine uracil and cytosine
Explain the three types of RNA - structure and brief function
Three types of RNA are involved in protein synthesis:
messenger RNA (mRNA)
➜ long, single stranded molecule
➜ synthesised in nucleus and carries DNA genetic code
to Ribosomes
➜ different mRNA molecules have different lengths depending
on genes from which they are synthesised.
ribosomal RNA (rRNA)
➜ found in cytoplasm
➜ comprises large complex molecules, ribosomes
➜ They are the site of translation of the genetic code.
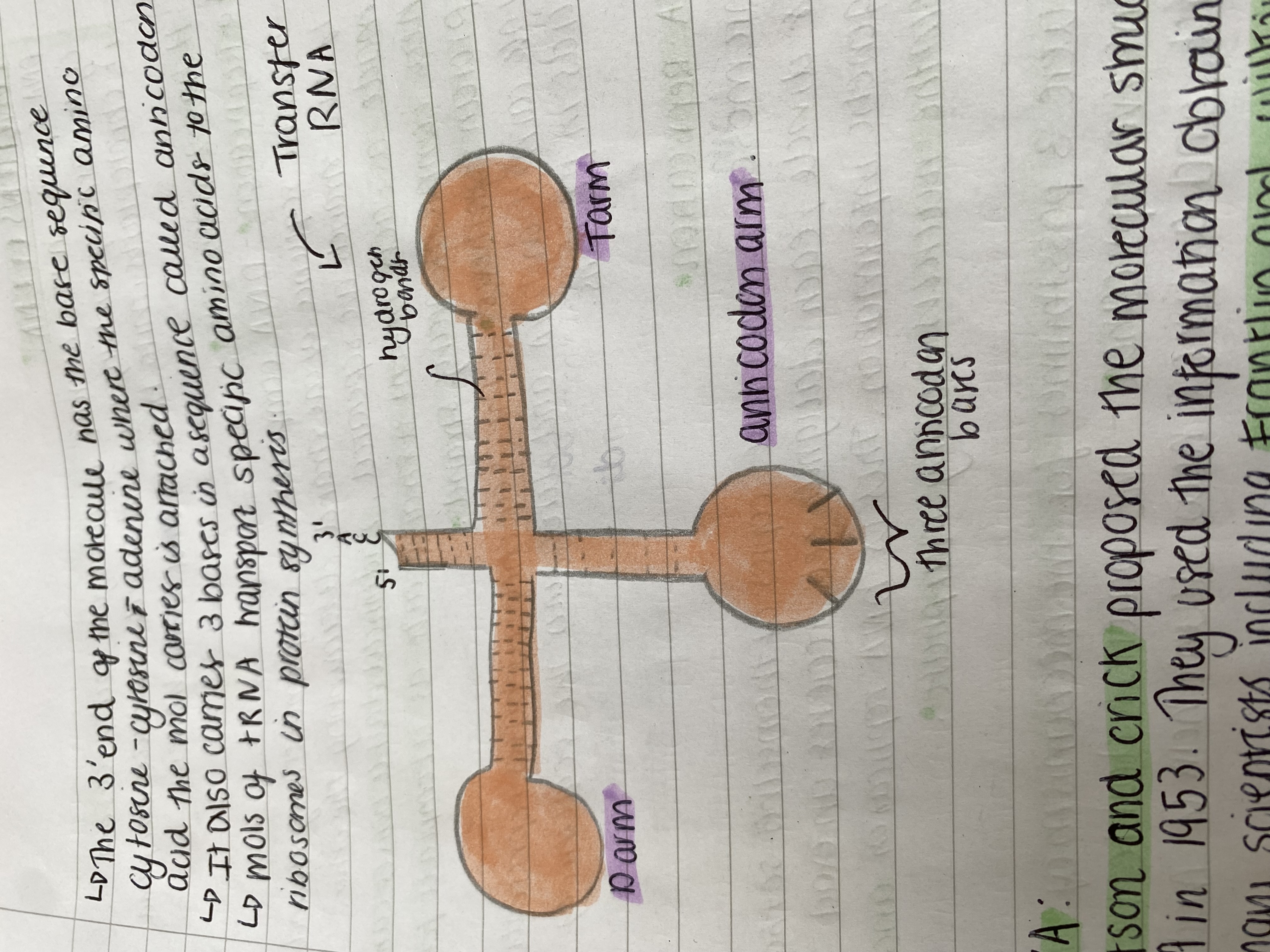
Transfer RNA (tRNA)
➜ small single stranded molecule
➜ it folds in places so there are base sequences forming complementary pairs.
➜ They have a clover leaf shape.
The 3' end of the molecule has the base sequence cytosine - cytosine - adenine where the specific amino acid the mol carries is attached.
It also carries 3 bases in a sequence called anticodon.
Mols of tRNA transport specific amino acids to the ribosomes in protein synthesis.
Explain briefly the roles of DNA
DNA is found in the nucleus of eukaryotic cells, loose in cytoplasm of prokaryotic and small molecules are found in chloroplasts mitochondria and some viruses.
It has 2 main roles:
Replication - DNA comprises two complementary strands, the base sequence of one strand determining the other. If two strands are separated, 2 identical double helices can be made as each parent strand acts as a template for the new complementary strand.
2. protein synthesis - The sequence of bases represents info carried on DNA and determines the sequence amino acids in proteins.
Explain the 1st role of DNA
DNA replication
Chromosomes must make copies of themselves so that when cells divide each daughter cell receives an exact copy of the genetic information. This is DNA replication and takes place in the nucleus during interphase.
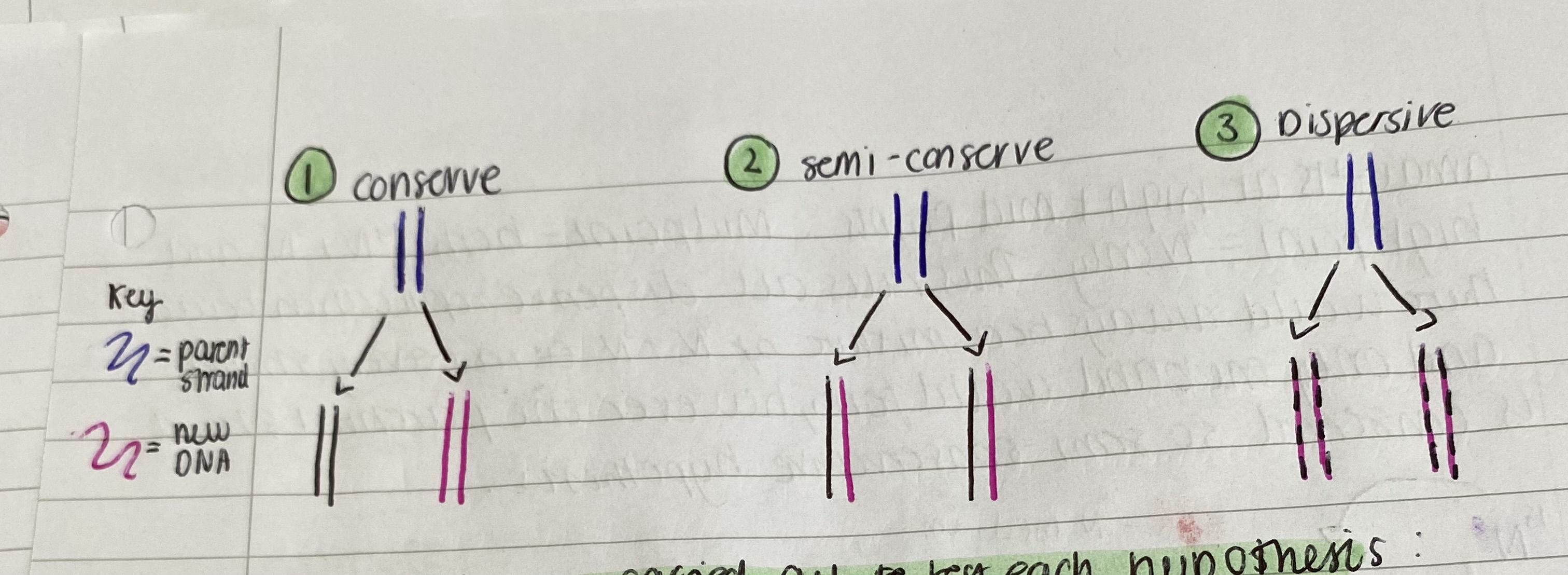
There were 3 possibilities for the mechanism of DNA
conservative replication - the parental double helix remains intact and a whole new double helix is made
semi-conservative replication - each parental double helix strand acts as a template once separated for the synthesis of the new strand -> each containing an original parental strand and a new synthesised complementary daughter strand.
Dispersive replication in which 2 new double helices contain fragments from both strands of parental double helix.
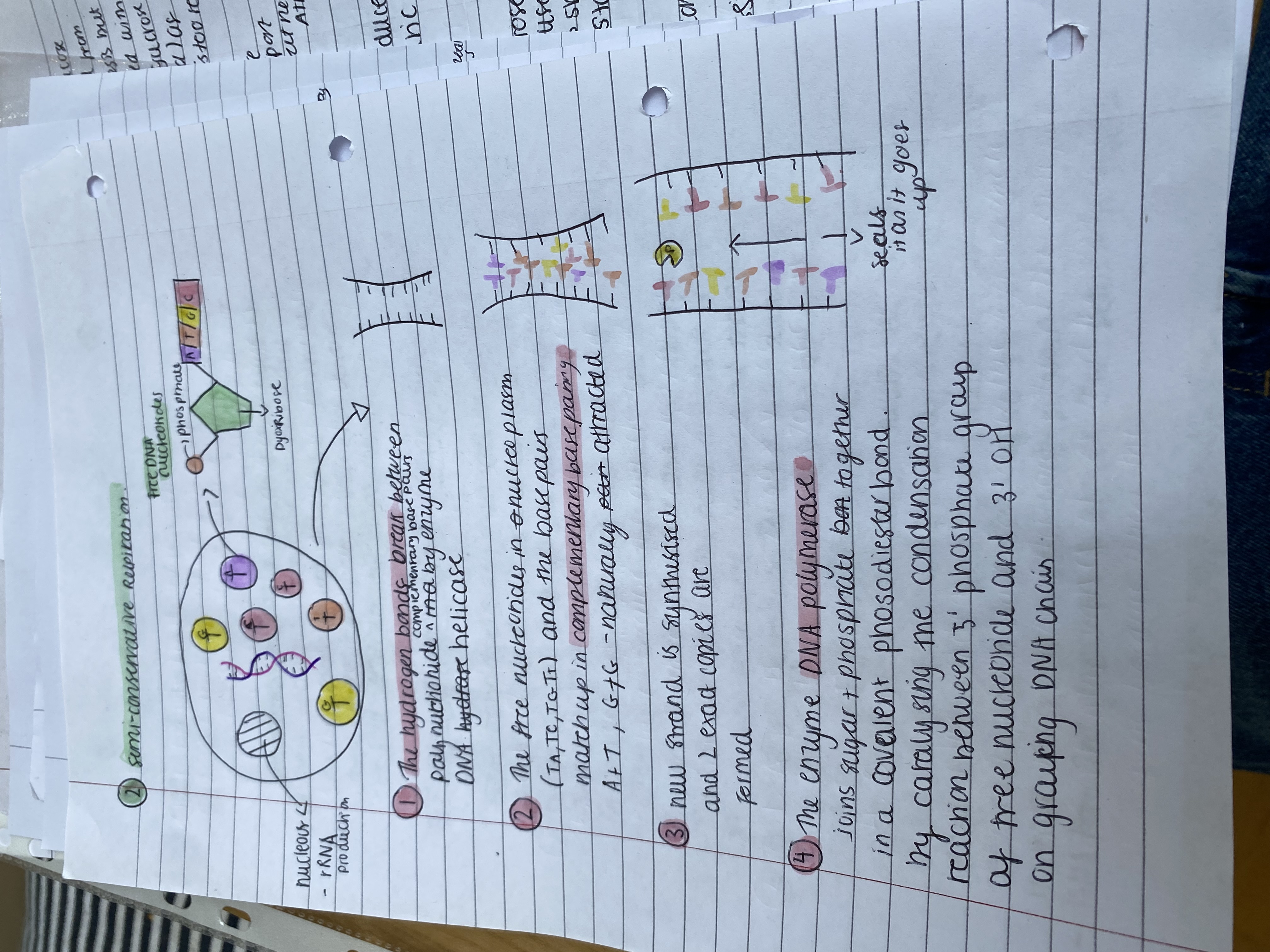
Explain the steps of semi conservative replication of DNA
semi-conservative replication
The hydrogen bonds break between polynucleotide strands by enzyme DNA helicase
The free nucleotides in nucleoplasm (T, A, T, C, T, G, T) and the base pairs match up in complementary base pairing A + T, G + C - naturally gets attracted
new strand is synthesised and 2 exact copies are formed
The enzyme DNA polymerase joins sugar + phosphate bond together in a covalent phosodiester bond by catalysing the condensation reaction between 5' phosphate group of free nucleotide and 3' OH on growing DNA chain.
Explain the 1st experiment on determining the type of DNA replication
Experiments were carried out to test each hypothesis
When Watson & Crick built their DNA model they realised that the complementary base pairs implied the two strands had separated, they would each made a new complementary strand - semi conservative replication.
Explain the 2nd experiment on determining the type of DNA replication
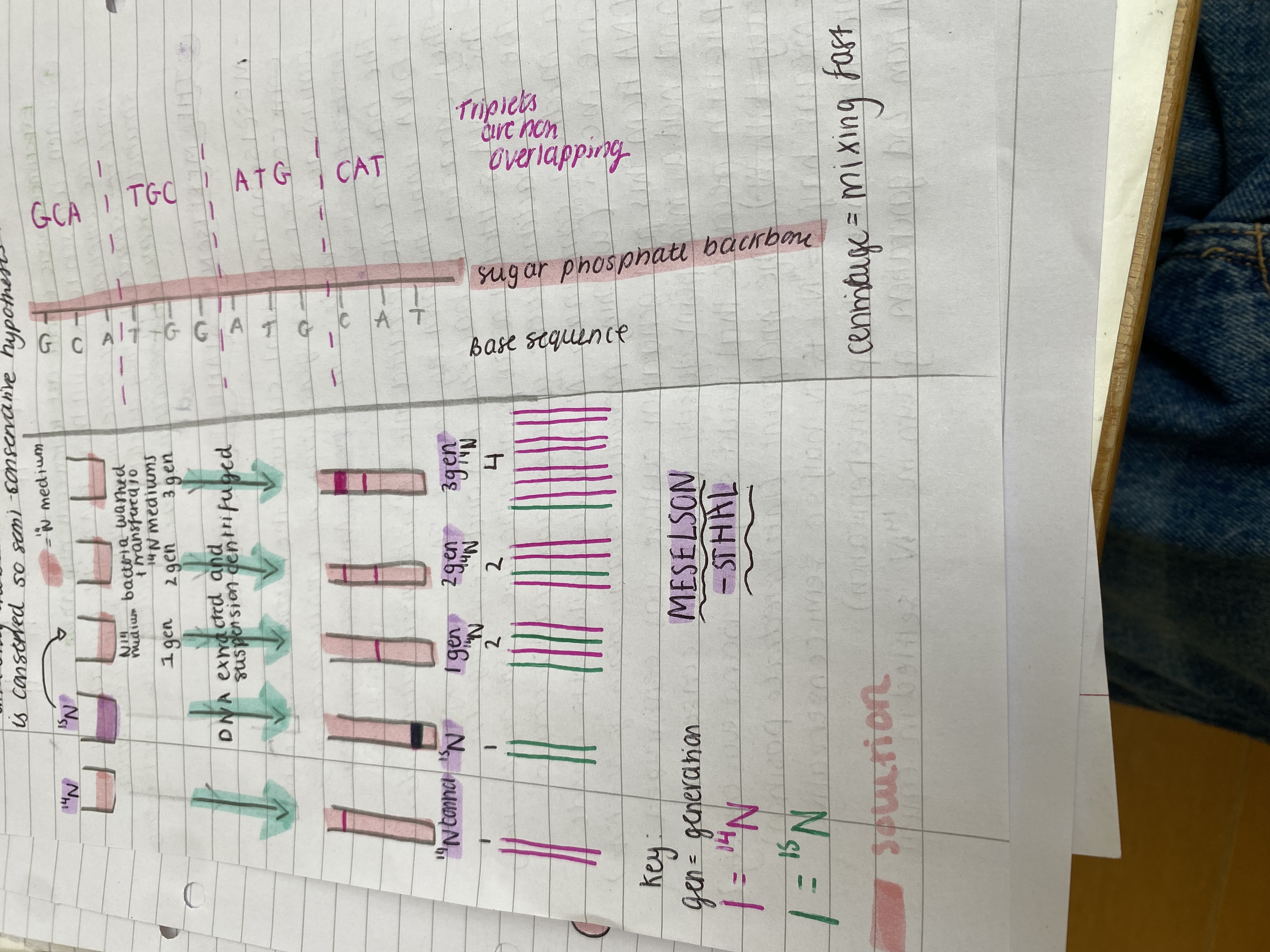
THE MESELSON - STAHL EXPERIMENT
Meselson and Stahl cultured the bacteria Escherichia coli for several generations in a medium containing amino acids made with a heavy Nitrogen isotope 15N instead of 14N.
The bacteria incorporated the 15N into their nucleotides + DNA until the DNA only contained 15N.
They extracted the bacteria DNA and centrifuged it.
The DNA settled at a low point in the tube (15N made it heavy).
The 15N bacteria were washed and transferred to a medium containing 14N and were allowed to divide once more. This washing prevented contamination of 14N + 15N.
DNA from this first generation culture was centrifuged and had a mid point density (ruled out conservative replication because that would say the band was entirely heavy with 15N). The mid point (intermediate position) means half 15N + 14N is 15N half so semi conservative replication or mixture (dispersive replication).
DNA from 2nd generation grown in 14N settled in equal amounts at high + mid points. Midpoint = both 14N + 15N and high point = 14N only. This rules out dispersive replication because there would always be a mixture of 14N + 15N in every strand and only one band would form, however one parental strand is conserved so semi-conservative hypothesis.
What’s the genetic code
DNA is the store of genetic information in the sequence of bases in the DNA in small sections of genes.
The base sequence directs the sequence of amino acids and therefore it determines the proteins like enzymes synthesized (enzymes are proteins so this determines reactions that take place in an organism).
What did experiments find out about the genetic code
The genetic code is a triplet code and experiments found...
Biochemical experiments showed that a polynucleotide strand always has 3 times the number of bases than an amino acid chain it coded for.
If 3 bases were removed from the polynucleotide chain, the polypeptide made would have 1 fewer amino acid and if the polynucleotide had 3 extra bases, the polypeptide would have 1x more amino acids.
These experiments concluded that 3 bases code for 1 amino acid.
This is supported by logical arithmetic: there are 4 different bases in DNA but over 20 different amino acids occur in proteins. This is because if 1 amino base pair coded for one amino acid then there would only be 4 types of amino acids (adenine, cytosine, thymine, guanine). If also 2 bases = 4^2 = 16 but 4^3 = 64 combinations > 20.
Explain the characteristics of the genetic code
Characteristics of the genetic code:
3 bases encode each amino acid so the genetic code is a triplet code.
64 combinations are possible but only 20 are found in proteins so more than one triplet can encode each amino acid so the code is 'degenerate' or 'redundant'.
The code is non-overlapping: each base occurs in only one triplet. So more than one codon for each amino acid
The code is punctuated so there are three triplet codes that don't code for amino acids. In mRNA they are called 'stop' codons and mark the end of a portion to the translation (eg. full stop).
The code is universal: the same triplet codes for the same amino acid in all known organisms.
The code is sometimes quoted 'DNA triplets' or 'RNA codons'
They are complementary to each other eg. DNA triplet for the amino acid glycine CCC and the RNA codon coding for glycine is GGG.
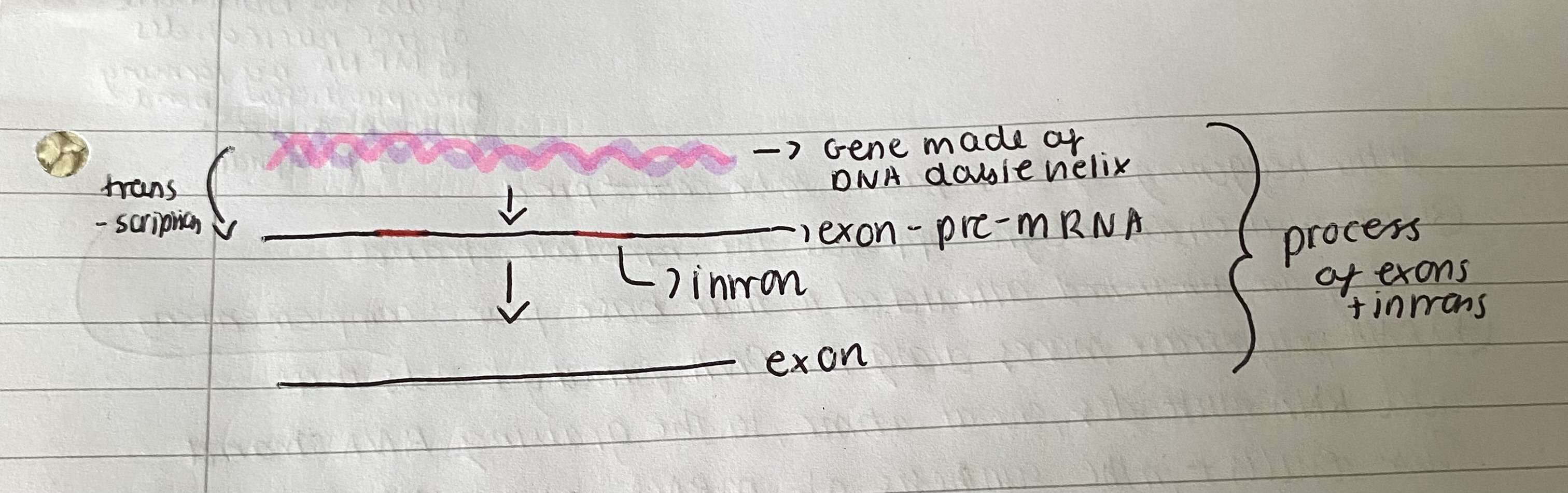
What are exons and introns and explain their significance
Exons and introns:
DNA contains the information for making polypeptides and an RNA version of the code is first made from DNA.
In prokaryotes the mRNA directs the synthesis of the polypeptide.
In eukaryotes, the RNA has to be processed in order to be used in synthesis of polypeptide - one gene, one polypeptide theory is incorrect. The introns are removed in the nucleus.
In eukaryotic organisms the initial RNA version of the code is longer than the mRNA as it contains base sequences that need to be removed. This is sometimes called 'pre-messenger RNA' (pre-mRNA). They are cut out because it could wouldn't form the protein.
The sequences that are removed are introns. They are not translated into proteins as they are cut out by endonucleases. The endonucleases cut out introns with DNA as substrate.
The sequences left are the exons which are joined together or spliced with ligases to form mature RNA.
If not removing introns the polypeptide chain that forms that specific protein won't be formed so the protein won't be formed - bad effects.
What are the two stages of protein synthesis - into
PROTEIN SYNTHESIS:
Here are the stages of protein synthesis:
Transcription - one strand of DNA acts as a template for production of synthesis of mRNA which is a complementary section of the DNA sequence. This then directs the synthesis of a complementary sequence of RNA too with the enzyme polymerase. It occurs in nucleus.
Translation: The sequence of codons on the mRNA acts as a template to which tRNA molecules attach and the amino acids they carry are assembled into a polypeptide of a specific sequence of amino acids at ribosomes.
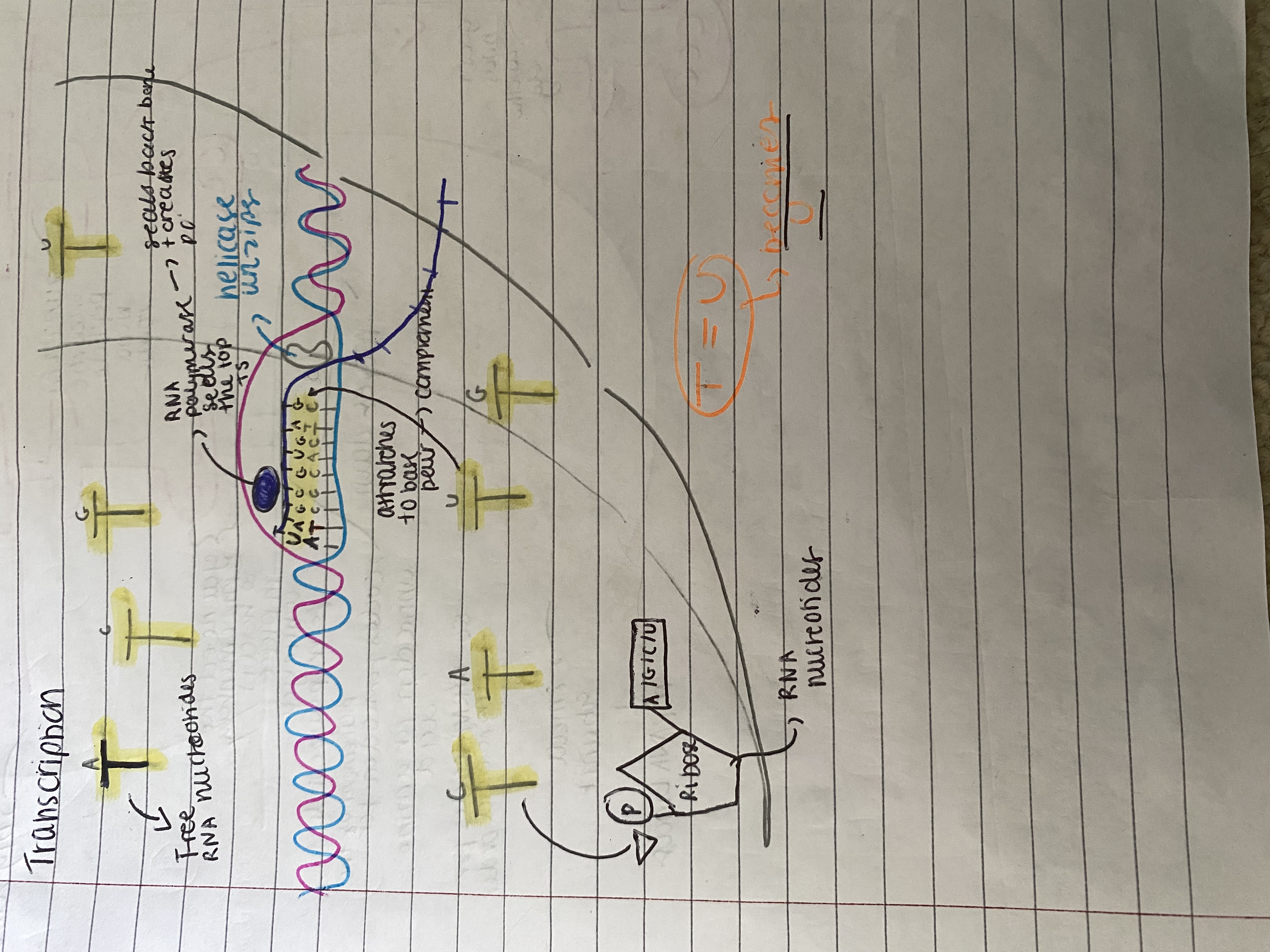
Explain in detail transcription and its stages
Transcription
The DNA doesn't leave the nucleus but instead DNA gene acts as templates for a mRNA production which carries the information needed for protein synthesis (order of amino acids in primary sequence) from the nucleus to ribosomes in the cytoplasm. The ribosomes provide a suitable surface for the attachment of mRNA and assembly of protein.
Steps of transcription:
DNA helicase enzyme breaks hydrogen bonds between complementary bases in a specific region of the DNA molecule. This causes 2 strands to separate and unwind exposing nucleotide bases.
enzyme RNA polymerase binds to the template strand of DNA At the beginning of the sequence to be copied,
free nucleotides align opposite the template strand as they are attracted to their base pair - complementary.
RNA polymerase moves along the DNA forming bonds that add RNA nucleotides, one at a time, to the growing RNA strand - seals the backbone of free nucleotides to mRNA by forming phosphodiester bond between ribose + phosphate. This results in the synthesis of mRNA along the portion of unwound DNA. Behind RNA polymerase, the DNA strands rewind to reform the double helix.
The RNA polymerase separates from the template strand when it reaches a 'stop' signal.
The transcript is complete and the mRNA detaches from DNA.
Explain translation and the steps in detail
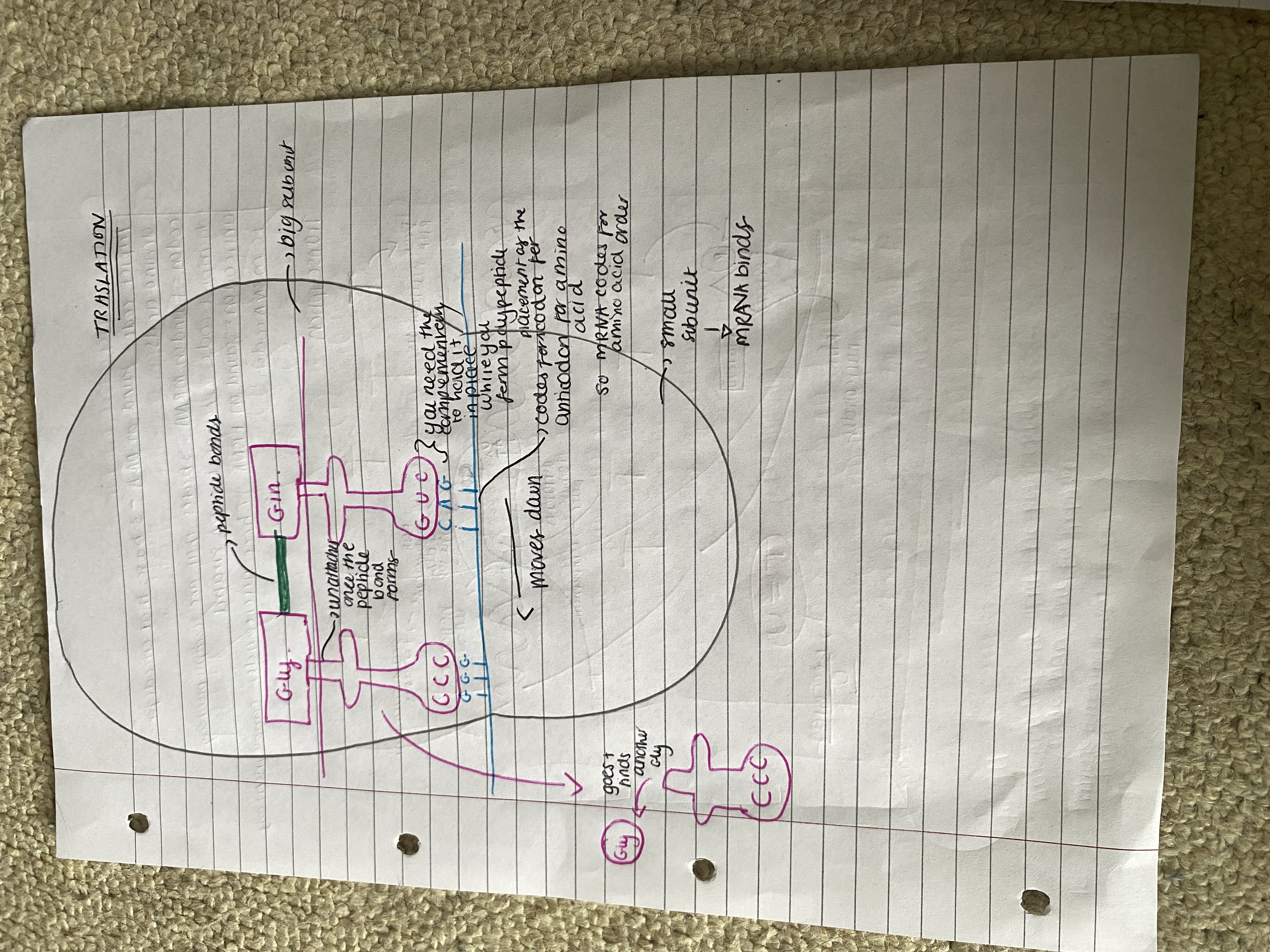
Translation:
In translation, the sequence of codons on mRNA is used to generate a specific sequence of amino acids - forms a polypeptide. Takes place on ribosomes & uses tRNA. Each ribosome has 2 subunits involved in translation:
The larger subunit has 2 attachment sites for tRNA so 2 tRNA are associated with each ribosome
The smaller subunit binds to mRNA
The ribosome acts as a framework moving along the mRNA and holding the codon-anticodon complex together until the 2 amino acids attached to adjacent tRNA molecules bind.
The ribosome moves along the mRNA adding one amino at a time until the polypeptide forms. (The order of bases in mRNA has determined the order of amino acids.)
Steps in translation:
Initiation: a ribosome attaches to a 'start' codon at one end of mRNA
The first tRNA with an anticodon complementary to the first codon on mRNA attaches to ribosome. The three bases at the codon on mRNA bond to 3 anticodon complementary bases with hydrogen bonds.
The same occurs for the second tRNA binding to complementary to 2nd codon with an anticodon and attaches to 2nd attachment site and codon + anticodon bond with hydrogen bonds.
Elongation: The two amino acids are sufficiently close for a ribosomal enzyme to catalyse a peptide bond between them.
The 1st tRNA leaves the ribosome so the attachment site is vacant. It returns to cytoplasm to bind to another copy of its specific amino acid.
The ribosome moves one codon along the mRNA strand
The next tRNA binds
Termination: the sequence repeats until a 'stop' codon is reached
The ribosome-mRNA-polypeptide complex separates usually several ribosomes bind to one mRNA strand all reading the coding at the same time. POLYSOME - several polypeptides can be made.
What is the difference between a codon, anticodon and triplet code
Triplet code: found on DNA - 3 bases that encode for amino acids
codon: found on mRNA - 3 bases that are complementary to triplet code to template DNA strand
anticodon: found on tRNA. Three nucleotides complementary to mRNA codon.
What is tRNA and amino acid activation
TRNA and amino acid activation:
Once tRNA is released from the ribosome it is free to collect another amino acid from the amino acid pool in cytoplasm.
Energy from ATP is needed to attach the amino acid to tRNA - This is amino acid activation. And an enzyme is used
What is the development of the one gene one polypeptide hypothesis
Gene + Polypeptide
After an understanding of genetic material was made from DNA was reached, they wondered how the information for the whole organism could be encoded.
Experiments on fungus Neurospora crassa in the 1940's found that radiation damage to DNA prevented any enzyme (protein) being synthesized.
This lead to the one gene-one enzyme hypothesis (one protein).
(alpha) (beta) (x2) genes needed. very diverse biological
However, it was then found that haemoglobin and many other proteins have more than one polypeptide chain due to structure
so one gene - one polypeptide was developed as a hypothesis.
It’s not actually one gene though because you can get more than one polypeptide for each gene due to removing differnet introns to form different polypeptides for differnet proteins
Why the one gene one polypeptide hypothesis isn’t correct
The reason why the one gene hypothesis is incorrect is because one gene can code for more than one polypeptide. Exons can be spliced in different ways to remove different introns to form different polypeptide chains.
The other reason this theory doesn't work is due to post translational modification. You can add different functional groups (in golgi) to form different proteins so one gene ≠ polypeptide.
What is post-translational modification
Post translational modification:
The base sequence of a gene determines the primary structure of the polypeptide.
Proteins then have many functions in living organisms eg. enzymes, antibodies, structural, transport proteins so the proteins are modified at Golgi body.
Sometimes the primary structure itself is functional but mostly it is folded into secondary, tertiary, quaternary structures etc. and is modified.
adding carbohydrates -> glycoproteins
adding lipids makes -> lipoproteins
adding phosphate -> phospho-proteins