bio u3o1 (nucleic acids, proteins, DNA manipulation)
1/101
Earn XP
Description and Tags
dna stuff
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
102 Terms
what is a vector and why are recombinant plasmids referred to as such?
Vectors are agents that deliver foreign DNA sequences from one organism to another. In gene cloning, recombinant plasmids are inserted with foreign DNA, which would be carried into a bacterium, where the DNA from the plasmid is replicated into the bacterium.
Monomer
the smallest component or subunit of each organic molecule.
Polymer
formed when monomers join together
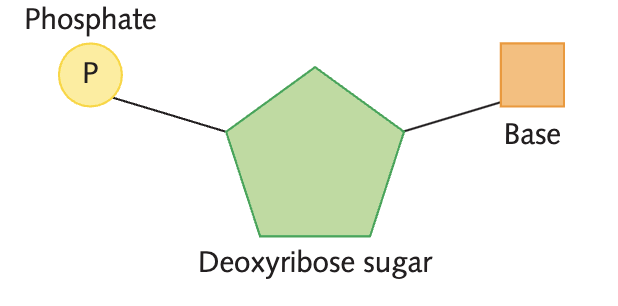
3 components of a nucleotide
a five-carbon pentose sugar (either ribose or deoxyribose)
a negatively charged phosphate group
an organic nitrogen-containing compound called a base
Nucleic acid are…
Biomolecules containing the genetic code of every living organism is contained in its DNA
Draw the 3 components of the nucleotide (DNA and RNA)
deoxyribose for DNA
ribose for RNA
What type of bond do nucleotides have connecting the sugar-phosphate backbone
Strong covalent phosphodiester bonds
What differentiates RNA from DNA
single stranded
uracil instead of thymine
ribose sugar instead of deoxyribose sugar
much shorter (base pairs) than DNA
explain the functions of the 3 types of RNA
Messenger RNA (mRNA) transcript copy of a specific DNA sequence (encodes for synthesis of polypeptide)
Transfer RNA (tRNA) carries the amino acids to ribosomes for polypeptide synthesis
○ One end contains 3 bases that make up an anticodon
○ Other end is a region that attaches to one specific amino acid
Ribosomal RNA (rRNA) a primary component of the ribosome, responsible for its catalytic activity
Detail the function and location of mRNA
Located in the nucleus and cytosol
mRNA carries a transcript copy of a specific DNA sequence that encodes for the synthesis of polypeptides.
carries the DNA code from the nucleus to the ribosome
Detail the function and location of tRNA
tRNA molecules each carry and then deliver one specific amino acid of the 20 amino acids
Three unpaired nucleotides at one end of the tRNA molecule are called the anticodon, which binds to a complementary mRNA codon (base-pairing rules)
amino acid is attached to an amino acid acceptor site on the stem of the tRNA molecule
free-floating molecules within the
cytosol
Detail the function and location of rRNA
found in cytosol
primary component of the ribosome
associates with proteins to form ribosomes
Compare uracil and thymine
Thymine is found in DNA whilst uracil is found in RNA
condensation polymerisation
a reaction in which monomers are linked together into a polymer with the release of a small molecule, such as water, as a by-product. The reaction that forms the peptide bonds joining adjacent amino acids.
Purine bases
Adenine and guanine
They are larger and have 2 rings made of carbon and nitrogen
Pyrimidine bases
Cytosine, Uracil, Thymine
They are smaller and have 1 six-membered ring containing carbon and nitrogen
genetic code
a universal triplet code that is degenerate and the steps in gene expression, including transcription, RNA processing in eukaryotic cells and translation by ribosomes
what are genes and how does it code for protein?
Genes are a segment of DNA in a chromosome that codes for a polypeptide, comprised of the prompter, exons, and introns. The code is written in codons, a set of 3 nucleotides corresponding to an amino acid. It is the chain of amino acids in certain orders that creates the proteins.
Degenerate/redundant in reference of genetic code and its significance
A property of the genetic code where two or more codons encode most amino acids.
This means that if a mutation occurs in the DNA, there is less chance that it will result in an amino acid change when the codons are read, meaning that the resulting protein can still be functional.
The term ‘universal’ in reference of the genetic code
All organisms use the same sequence of codons to code for the same 20 amino acids.
3 steps of transcription
INITIATION
• RNA polymerase binds to specific promoter sequence in upstream region of DNA strand
• Triggers an unwinding and separation of DNA strands, exposing bases of template strand
2. ELONGATION
• RNA polymerase moves along coding sequence in 5’ → 3’ direction
• Free RNA nucleotides align opposite to their complementary base partner.
• New nucleotides are added onto the growing mRNA strand
• RNA polymerase covalently bonds RNA nucleotides together
3. TERMINATION
• RNA polymerase moves into downstream region
• At the terminator (polyadenylation sequence for eukaryotic cells), both RNA polymerase and the newly formed pre-mRNA strand detach from the DNA template.
• DNA rewinds
Molecules involved in RNA transcription
RNA polymerase
transcription factors
pre-RNA is synthesised
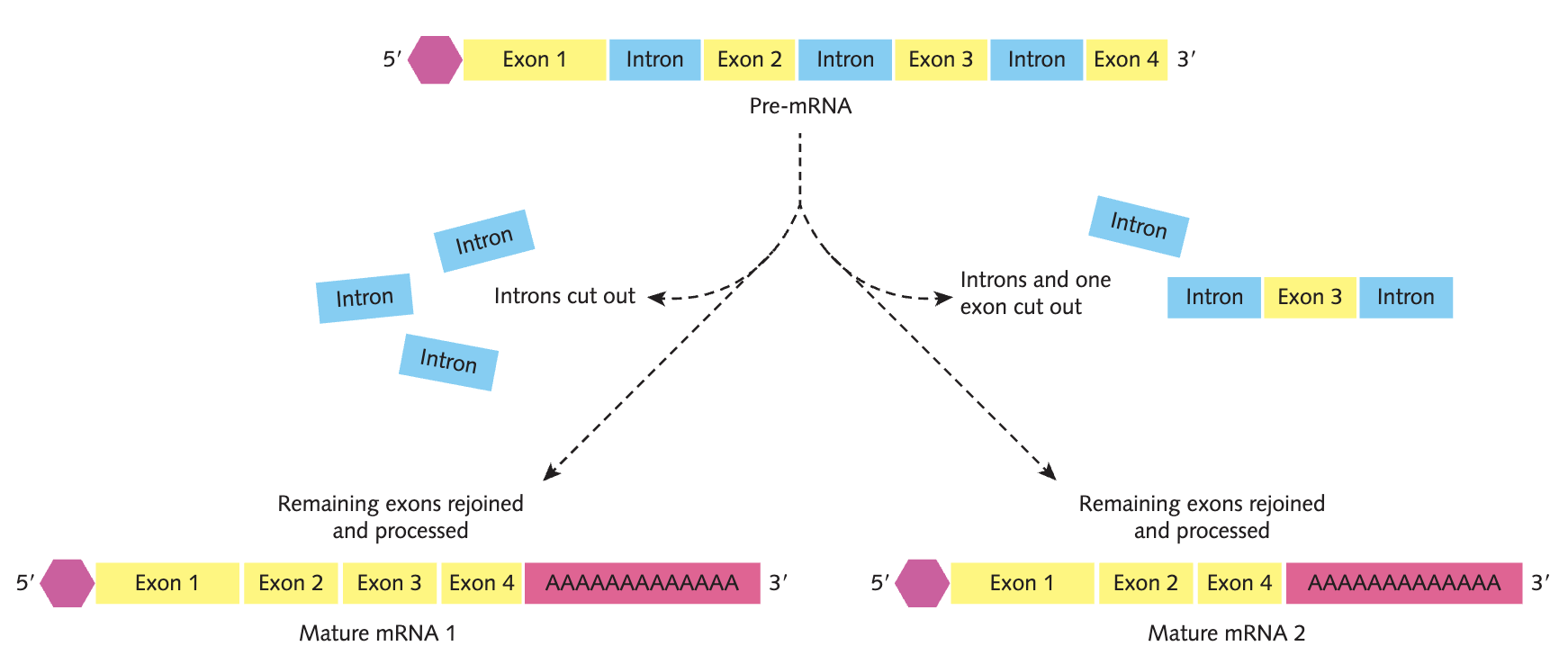
explain RNA processing in terms of introns and exons
In RNA processing, introns, the non-coding regions of the pre-mRNA, are spliced out (by spliceosomes), and exons, regions containing the code of amino acids, will be kept.
the purpose of each of the post-transcriptional modifications to eukaryotic pre-mRNA
Exon splicing
> so the mRNA can encode a protein with the right sequence
5’ cap
> methyl group is added to 5' end of transcript to protect against degradation by exonuclease
> aid ribosomes to attach to the 5′ end of the mRNA once the mRNA reaches the cytoplasm.
3’ Poly A tail
> with up to 250 adenines added to 3' end to increase stability of mRNA and facilitate mRNA export from nucleus
alternative splicing and its importance
RNA processing removes some exons along with the introns, producing mRNA molecules with different lengths and sequences from the same pre-mRNA sequence. Alternative splicing produces different polypeptides from the same genes, increasing the number of different protein products.
Key events in translation
Initiation
mRNA is brought to ribosome and is read in groups of 3 (codons)
Translation begins at AUG (start) codon
Elongation
tRNA brings specific amino acids according to mRNA sequence and aligns anticodons with mRNA codons
Ribosomes move along mRNA strand and facilitate peptide bonding between amino acids, resulting in polypeptide structure
Termination
A stop codon brings in a release factor that breaks the emerging polypeptide free while the mRNA and ribosome dissociate.
Molecules involved in translation
mRNA
tRNA
ribosome/rRNA
promoter region’s significance in transcription
The promoter region signals the start of the gene. Transcription begins when transcription factors position the RNA polymerase onto the DNA to bind with the promoter region.
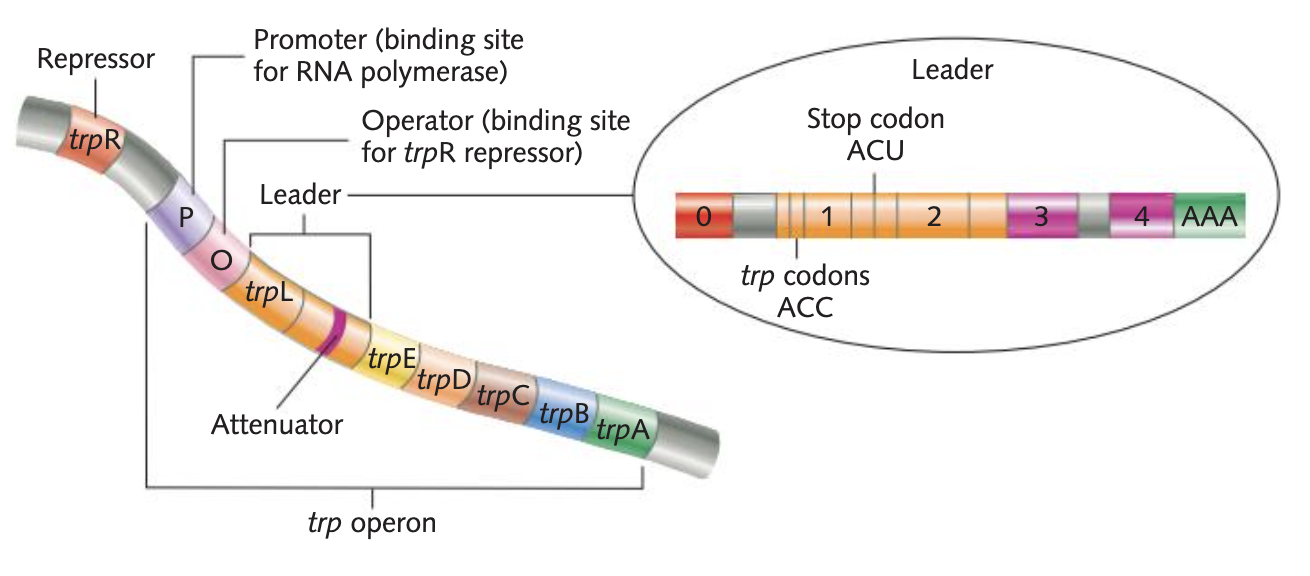
What comprises the basic structure of a eukaryotic gene
Promoter: the segment of DNA where RNA polymerase binds to begin transcription
Operator: where repressor molecule binds to operon model, preventing transcription from occurring
Upstream: region towards the 5' end before the coding region, could include the TATA box
Coding region: the part of a gene that contains the information that codes for a protein
regions next to the coding regions are called flanking regions
has introns (non-coding) and exons (coding)
Downstream: towards the 3' end before the coding region, could include the polyadenylation (AAUAA) sequence
Promoter
A segment of DNA where the RNA polymerase binds to begin transcription. The promoter does not code for genes but signals the start of the gene.
Operator
A segment of DNA where a protein binds, usually to switch off gene exp
Structural genes
Genes producing protein products that become part of the structure and functioning of the organism.
A gene that codes for any RNA or protein product other than a regulator
e.g. tRNA, rRNA, enzymes
Regulatory gene
A gene whose production switches on or off the expression of one or more genes.
A gene that codes for a product (generally protein) that controls the expression of other genes.
Positioned at the end of the promoter region but before the coding regions
e.g repressor protein
Repressor protein
A product of a regulatory gene that is located away from the operon it acts on and has its own promoter.
They are specific for the operator of the operon they bind.
When bound, transcription cannot occur
Therefore affects gene expression
Operon
A group of structural genes expressed as a single unit. Includes the promoter, the operator, and the leader segment.
Pneumonic: OPOGS!
Operator: Promoter, Operator, Gene (Structural)
purpose of the trp structural gene (Trp E/D/C/B/A)
trpA, trpB, trpC, trpD and trpE are the 5 structural genes that code for the enzymes that produce tryptophan
identify the promoter, leader, attenuator, terminator, regulator and operator regions of the trp operon
transcription factors role in transcription
transcription factors control the rate of transcription by binding to a specific DNA sequence. Transcription factors can serve to initiate or enhance transcription, or they may act to prevent it.
how can repressor proteins prevent transcription
Repressor protein requires tryptophan in order to be active.
• Tryptophan binds to a repressor, triggering a conformational change that activates the protein
• When tryptophan levels are high, trp repressor is active and operon is switched off
• When tryptophan levels are low, trp repressor is inactive and the operon is switched on
Trp repressor binds to an operator site and inhibits expression by preventing the binding of RNA polymerase to promoter
explain how the tryptophan molecule can inhibit/repress the trp operon into being expressed
When there is NO/LOW TRYPTOPHAN
the repressor protein is inactive and is unable to bind to the trp operator
allows RNA polymerase to transcribe trp genes
When tryptophan is PRESENT or high
repressor is active and binds to trp operator
prevents transcription from occurring
Tryptophan is a corepressor in this case.
In attenuation,
When there is high tryptophan, the ribosome travels at a faster speed, blocking the formation of the anti-termination loop and dissociating RNA polymerase from DNA, stopping transcription.
When there is low tryptophan, ribosomes stall due to the absence of trp, allowing for the formation of an anti-termination loop; therefore, transcription continues.
how can attenuation regulate the expression of the Trp operon
When there is high tryptophan, the ribosome travels at a faster speed, blocking the formation of the anti-termination loop and dissociating RNA polymerase from DNA, stopping transcription.
When there is low tryptophan, ribosomes stall due to the absence of trp, allowing for the formation of an anti-termination loop; therefore, transcription continues.
What molecule is released in the condensation polymerisation reaction?
water molecule
condensation polymerisation
a reaction in which monomers are linked together into a polymer with the release of a small molecule, such as water, as a by-product
The bond between two adjacent amino acids is
a peptide bond is the result of the condensation polymerisation reaction; it bonds two amino acids in a chain
Primary structure of protein folding and its bond
The primary structure is the linear sequence of amino acids that makes up the polypeptide chain. The amino acids are linked with a peptide bond.
Secondary structure of protein folding and its bond
The localised folding of the polypeptide chain is due to hydrogen bonding between peptide bonds of neighbouring amino acids. The coiled and folded portions of the polypeptide chain form the secondary structure. The tight coils are referred to as alpha-helixes, the flattened foldings are beta-pleats, and any remaining arrangements are random coils.
Tertiary structure of protein folding and its bond
The overall 3-dimensional shape of a completely folded polypeptide.The interactions between R groups of amino acids (hydrophilic/hydrophobic) cause the polypeptide chain to be folded and twisted into the protein’s conformation.
the tertiary structure determines the biological functionality of a protein depending on the way the protein folds into shape
Quarternary structure of protein folding and its bond
The quaternary is formed when two or more polypeptides associate into the mature protein. A variety of hydrogen bonds, ionic bonds, and covalent bonds hold the polypeptide chains together in their final shape.
The significance of alpha-helixes and beta-pleats
They are important H-bonded secondary structures that provide strength and elasticity
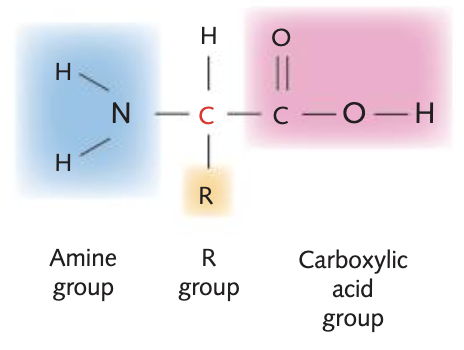
draw and label an amino acid
STRUCTURE OF AMINO ACIDS
central carbon atom
Amine (H2N)
Carboxyl group (COOH)
Variable R group
What causes a protein’s denaturation
Proteins may also lose their functional shape if they are:
exposed to high temperatures
in concentrated salt solutions
in very acidic or alkaline conditions
enzymes and its significance
a specific protein catalyst that increases the rate of biochemical reaction within a cell by lowering the amount of energy required for the reaction to proceed
proteome
The complete set of proteins expressed by a genome, cell, tissue, or organism at a certain time
proteomics
the study of proteomes
the role of ribosomes in the synthesis of proteins
The process of translation reads the mRNA code to produce a polypeptide chain that will undergo folding to become a mature protein. In translation, the ribosome serves as the workbench for protein synthesis, while tRNA molecules provide the raw materials. Therefore, ribosomes are crucial molecules in protein synthesis.
Where can ribosomes be found in a cell
embedded in the rough endoplasmic reticulum
cytosol
rough endoplasmic reticulum’s role in the protein secretory pathway
Proteins enter the lumen of the rough ER to be assisted to fold in the correct way.
Then they are sorted and transported to the golgi apparatus by transport vesicles.
golgi apparatus/body role in golgi apparatus/body
Proteins are modified by enzymes in the cisternae of the golgi apparatus to add or remove components until it reaches its mature, functional form. Then the proteins are packaged in secretory vesicles to transport them out of the cell.
does exocytosis use energy
well yes (and vesicles)
sequence of events involved in the protein secretory pathway
Nucleus → Rough ER → Golgi body → Vesicle
Synthesis of RNA at nucleus
Protein synthesis at ribosome
Rough ER assists in protein folding and then transports it to Golgi
Golgi modifies proteins into mature form, then packages them into secretory vesicles for export
Vesicle merges with plasma membrane to secrete protein out of cell via exocytosis
the role of polymerases (DNA, RNA and reverse transcriptase) in the synthesis of nucleotides
Polymerases are enzymes that catalyse the formation of DNA and RNA from nucleotide monomers.
taq polymerase
From a bacteria found in hot springs
optimal temperature of 72°C
used in PCR as it is stable at high temperatures
role of an endonuclease
restriction endonucleases/enzymes are the cutting tools that cleaves or cuts nucleic acid
where do restriction enzymes cut
Restriction enzymes only cut specific sequences of DNA (restriction sites/recognition sequences)
Most recognition sequences are palindromes of their complementary sequence.
Look for the 6-base palindromic sequence
blunt vs sticky ends
When restriction enzymes cut DNA, they can produce two types of cuts:
Sticky ends
Overhanging sections of exposed nucleotides
Blunt ends
Straight cut, no exposed nucleotides.
how are restriction enzymes used in genetic engineering
The bacterial DNA and the gene to be inserted into the bacterium are cut with the same restriction enzyme to ensure their ends match.
where endonucleases are found and the importance of this
endonucleases are naturally occuring in bacteria
they all cut different restriction sites
different bacterias will yield different endonuclease
a collection of bacteria is needed for their endonuclease to
significance of endonuclease in genetic engineering
When you wish to insert a piece of DNA into an existing DNA, you must first cut each DNA with the same restriction enzyme/endonuclease
Now the new piece of DNA is ready to insert, and DNA ligase is added to join the nucleotides together!
explain the role of ligase
DNA ligase is used to join fragments of DNA together. These enzymes will help to create the strong covalent bonds between the DNA nucleotides.
Helps to create the phosphodiester bond
Reassembles (glues) the DNA back into being double-stranded
phosphodiester bonds
In DNA nucleotides, the sugar-phosphate groups that make up the backbone of each strand are linked by phosphodiester bonds
what is a CRISPR locus, and how does it work to provide “immunity” for bacteria
The CRISPR loci are regions of a bacteria’s genome where small fragments of a virus/bacteriophage DNA, called protospacers, are inserted.
The CRISPR loci allow the bacteria to "remember" the viruses (or closely related ones).
Transcription of the CRISPR spacers and CRISPR palindrome repeats produces an RNA molecule called crRNA.
crRNA is complementary to the spacers
This crRNA binds to Cas9 proteins to produce a CRISPR-Cas9 complex. This complex then scans the bacterial cell for any future infecting bacteriophage DNA that is complementary to that in the crRNA. The Cas9 will only cut out the target sequence if it recognises a very short (2-6 nucleotides) sequence adjacent to the spacer called a PAM. If located, the Cas9 endonuclease proteins cleave the phosphate-sugar backbone of the bacteriophage DNA, inactivating and destroying it.
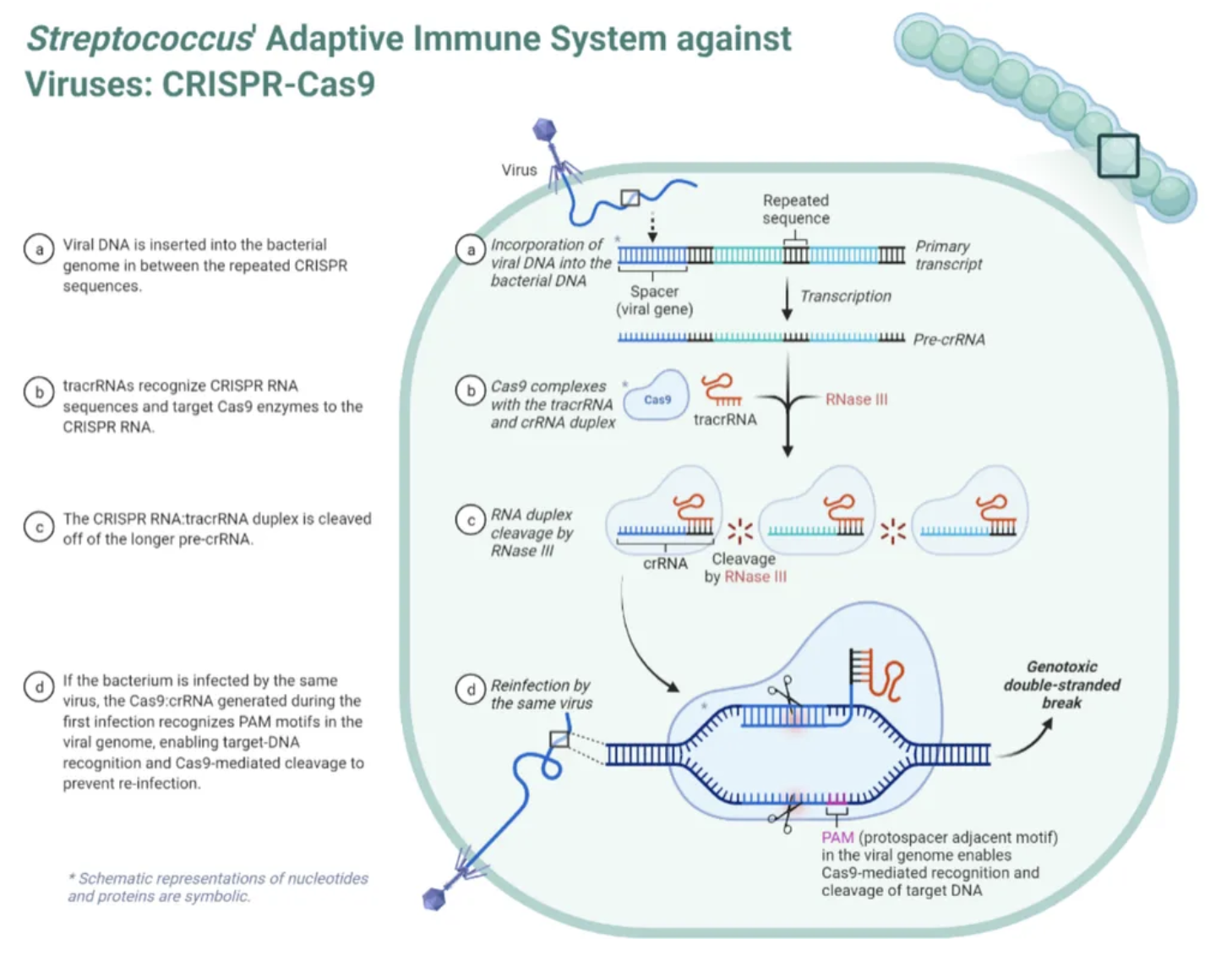
the role of the Cas9 enzyme
Bacterial immune system
The Cas9 proteins bind to crRNA to form the CRISPR-Cas9 complex. This complex scans for foreign DNA complementary to the crRNA, which the Cas9 will cut when detected.
Genetic engineering
In biotechnology, the bacteria-derived Cas9 enzyme is directed by a single guide RNA, which has a complementary target site in genomic DNA. The Cas9 will find the target PAM sequence, check whether the sgRNA aligns with the DNA, and then cut the selected sequence of DNA.
the sgRNA can be artificially constructed to guide where researchers want the Cas9 enzyme to cut
sgRNA
single guide RNA is the RNA that guides the Cas9 protein to the target sequence in a genome for gene editing.
how CRISPR-Cas9 genetic engineering can be used in a variety of applications
Since the CRISPR-Cas9 system is specific in where it cuts, DNA can be cut at any desired location.
There are numerous applications for CRISPR, from
altering immune systems to fight cancer
editing faulty alleles in diseases such as sickle-cell anaemia and cystic fibrosis
in agriculture: insertion or deletion of desired traits to improve yield or tolerance to environmental stress or longer shelf life
bioethical issues surrounding genetic technologies
Safety
>the possibility of off-target effects (edits in the wrong place) and mosaicism (when some cells carry the edit, but others do not
Informed Consent
> patients affected by the edits are the embryo and future generations
> consumers informed about consuming GMO foodsJustice and Equity
> only be accessible to the wealthy
Genome-Editing Research Involving Embryos
> as genetic material would be passed onto the next generation (if mutations arise)
> existing moral and religious objections
why do we use PCR and why we need to get multiple sequences of DNA
PCR stands for polymerase chain reactions. PCR is used to amplify or make multiple copies of a specific sequence of DNA. Genetic analysis and engineering both need enough copies of DNA to work effectively.
why is Taq polymerase used in the synthesis of nucleotides
It is stable at high temperature
> necessary as PCR requires high temperature to denature DNA strands
Taq polymerase synthesises new DNA strands in the extension stage
comes from bacteria
what are the main processes involved in a cycle of PCR
Denature
A DNA sample (target DNA) is obtained. It is denatured by heating at 95°C for 5 minutes. This breaks the hydrogen bonds between the bases (DNA strands are separated).
Annealing
The DNA is cooled to 50- 55°C. Primers are annealed (joined) to each DNA strand. The reduced temperatures allow for base pairings (formation of hydrogen bonds)
Extension
Heated back to 72°C, Taq polymerase binds to primers and moves along each strand, generating a complementary strand in the 5' to 3' direction, joining free nucleotides
applications of PCR
DNA sequencing via gel electrophoresis
DNA profiling in forensic science, determining family relationships and identifying human remains
the use of gel electrophoresis in relation to PCR
Gel electrophoresis separates fragments of DNA according to their size and charge. DNA is negatively charged, repelled by the negative electrode, and is sorted.
Know the general purpose of gel electrophoresis and its applications
Forensic and parental testing
negative and positive electrodes and their importance for the movement of DNA in gel electrophoresis
DNA is negatively charged and is repelled by the negative electrode. The power source is attached to the electrophoresis bath and switched on. Electric current causes negatively charged DNA to move through the gel towards the positive electrode. Smaller fragments move faster through the gel, moving further towards the positive electrode over a period of time.
How does size of DNA fragments determine their movement
Smaller strands (in terms of base pairs) can move faster than the larger strands. The position of bands on an agarose gel is used to determine the size of the separated DNA by comparing their location along the gel.
DNA profile + purpose
DNA profiling is the process where a specific DNA pattern, called a profile, is obtained from a person or sample of bodily tissue. Creating DNA profiles allows us to read fragments of DNA and analyse these responses via gel electrophoresis.
It is used in;
establishing paternity
solving crimes
Identifying human remains
diagnosing genetic disorders
how is DNA profile created
sample is obtained from a person
the DNA obtained is amplified by PCR
the DNA is sorted by gel electrophoresis
polymorphisms (genome regions of high variability between individuals) used to construct a DNA profile is called STRs
multiple STR loci are examined to obtain a profile
what is a plasmid and where it comes from
Small, extrachromosomal circular rings of DNA found in bacteria
why are plasmids used in gene cloning?
PCR DNA fragments do not survive long and are unstable
plasmids are copied many times in a bacteria cell and are copied when the bacteria cells replicate—this copies any DNA fragment inserted
plasmid’s circular structure makes it stable
plasmids are easily engineered to carry a number of different genes
Steps of gene cloning using plasmids
1. Plasmids are extracted from bacteria (via rupturing cell walls)
2. A restriction enzyme is used to cut the plasmid DNA at one point to create sticky or blunt ends, changing the plasmid from circular to linear
3. The same restriction enzyme is used to cut the “foreign DNA” of the gene so that it has sticky/blunt ends that it is complementary to the cut plasmid’s ends
4. The foreign DNA fragments and the plasmid is mixed
5. DNA ligase binds/glues the DNA to the plasmid. The DNA is now a permanent part of the recombinant plasmid after covalently bonding
6. The recombinant plasmids are added to the bacterial culture. They are taken up by some bacteria (transformation), in which they replicate.
7. Allow transformed bacteria to grow and then harvest and purify the cloned gene product.
bacterial transformation
a method of inserting a gene of interest into a plasmid (recombinant plasmid) and then transferring this into a bacteria.
steps in generating human insulin through recombinant plasmid
The human insulin gene* is cut from the chromosome using restriction endonuclease enzyme that produces sticky ends.
The plasmids are cut open using the same restriction endonuclease enzyme.
The human insulin gene is inserted into the plasmids using ligase enzyme.
The plasmid is inserted into the bacterial cell (transformation).
The transformed bacterial cell reproduces by binary fission.
The plasmid also independently replicates.
The gene product Insulin is collected and purified.
purpose of antibiotic resistance genes and reporter genes in a recombinant plasmid
Plasmid DNA contains genes for antibiotic (e.g. ampicillin) resistance
bacteria transformed is grown on medium supplemented with ampicillin
bacteria without the plamid will not grow as they do not have genes for ampicillin resistance
this process is called antibiotic selection
what is a GMO
GMOs are genetically modified organisms. For an organism to be identified as a GMO, any gene or DNA segment that is added or deleted through genetic engineering should be heritable.
transgenic organism
An organism whose genome has been artificially modified with DNA from a foreign species.
examples of GMO and TGO in agriculture that increase crop yield
Bt crops contain genes from bacteria that encode toxins harmful to a range of common crop pests.
AquAdvtage salmon is TGO, it has another fish’s promoter gene
Consequences-based
places central importance on the consideration of the consequences of an action (the ends), with the aim to achieve maximisation of positive outcomes and minimisation of negative effects.
Duty- and/or rule-based
concerned with how people act (the means) and places central importance on the idea that people have a duty to act in a particular way and/or that certain ethical rules must be followed, regardless of the consequences that may be produced.
Virtues-based
emphasises the individual goodness of the agent, and promotes acting in accordance with the values of a ‘moral’ person, such as honesty and compassion
Integrity
- the commitment to searching for knowledge and understanding and the honest reporting of all sources of information and communication of results.
- Integrity prioritises an accurate understanding and representation of the facts, whether favourable or unfavourable to an individual’s personal position, and encourages scrutiny and criticism.
Justice
justice is the commitment to fairness
Justice prioritises the fair distribution of resources, as well as equal access to the benefits of an action, policy, investigation, or research.
Beneficence
the commitment to maximising benefits and minimising the risks and harms involved in taking a particular position or course of action.