DNA: replication, transcription and translation.
1/71
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
72 Terms
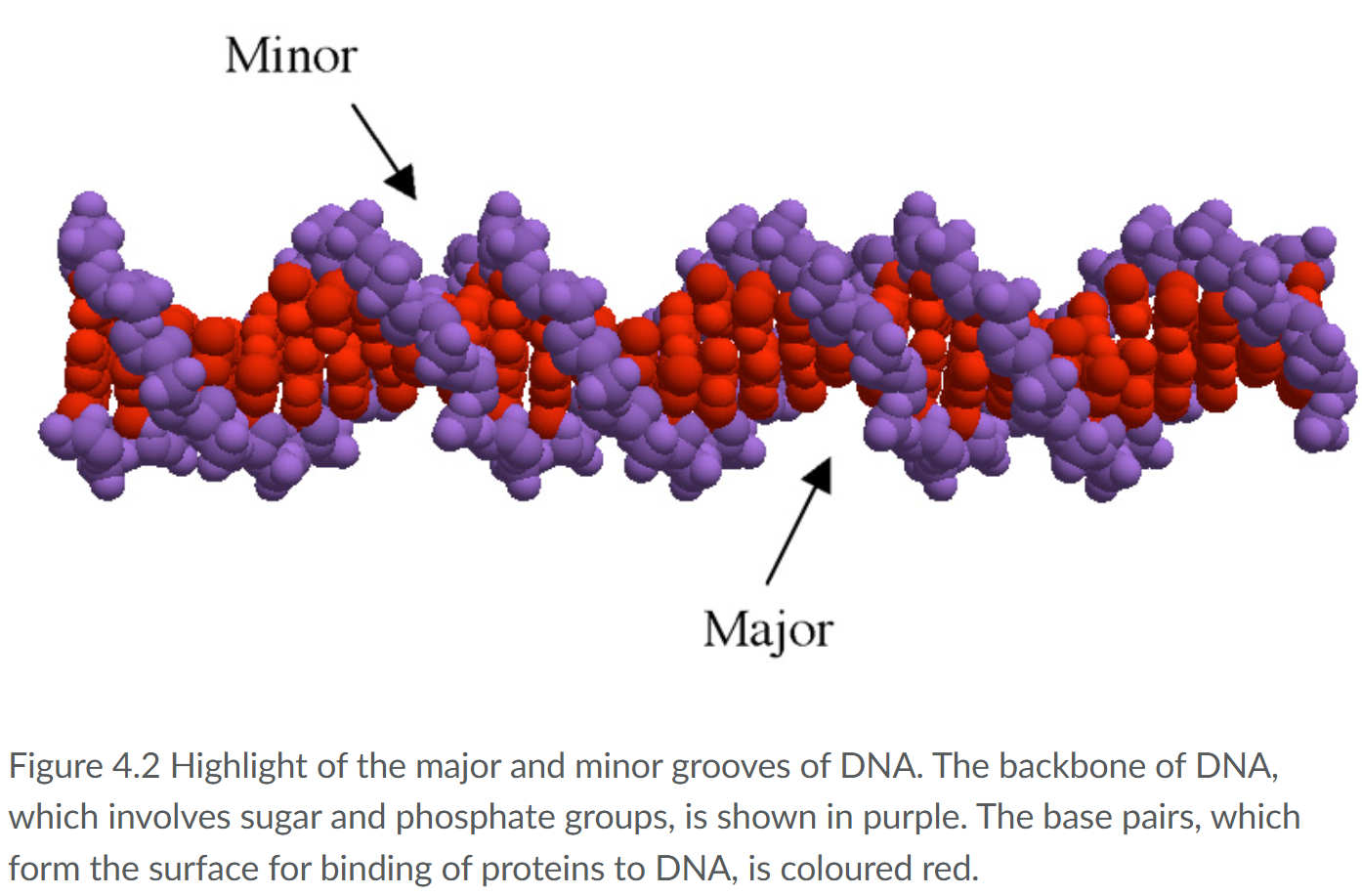
Which are the major and minor grooves of DNA?
The surfaces of these grooves are formed by the sides of the base pairs
this is relevant as for binding of DNA proteins as they hydrogen bond met the surfaces of the grooves.
What is the helix turn helix motif?
major structural motif capable of binding DNA.
-Each monomer incorporates two α helices, joined by a short strand of amino acids, that bind to the major groove of DNA.
- occurs in many proteins that regulate gene expression.
Dimer
2 protein molecule
tetramer
4 protien molecule
Nucleoside vs nucleotide
Nucleotide: sugar, base, phosphate
Nucleoside": the sugar and the base
What is the type of chain which DNA consists of called?
Polynucleotide
What is the model of DNA replication?
semiconservative
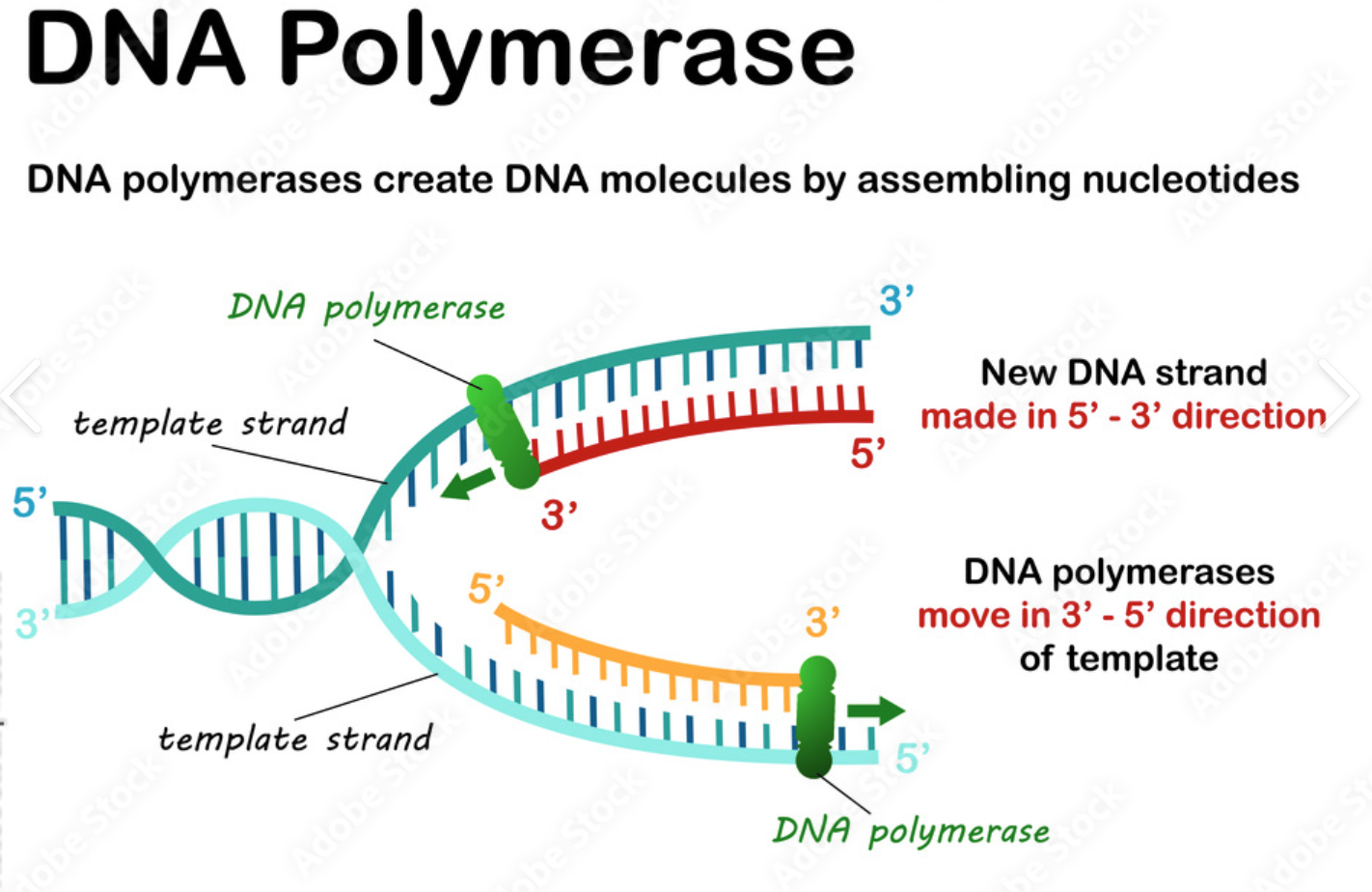
Function of DNA polymerase
ensures the correct base pair is formed
Catalyzes the synthesis of DNA in the 5' to 3' direction using a DNA strand as a template.
Nucleoside triphosphates base pairs met the base of the DNA template strand (3’) which forms a phosphodiester bond between the growing DNA and the new nucleotide.
van skills training:
DNA polymerase checks whether the correct nucleotide is bound to the DNA template before the nucleotide is covalently coupled to the newly synthesized DNA strand. In case a mistake is made (1 per 107 nucleotides), the enzyme uses “proofreading” activity.
During proofreading, the new DNA strand partially detaches van the template. DNA polymerase changes conformation, thus moving the wrong nucleotide van the polymerizing-site to the editing-site (error-correcting catalytic site). The wrongly inserted nucleotide is removed van the new DNA strand by a nuclease in the editing-site.
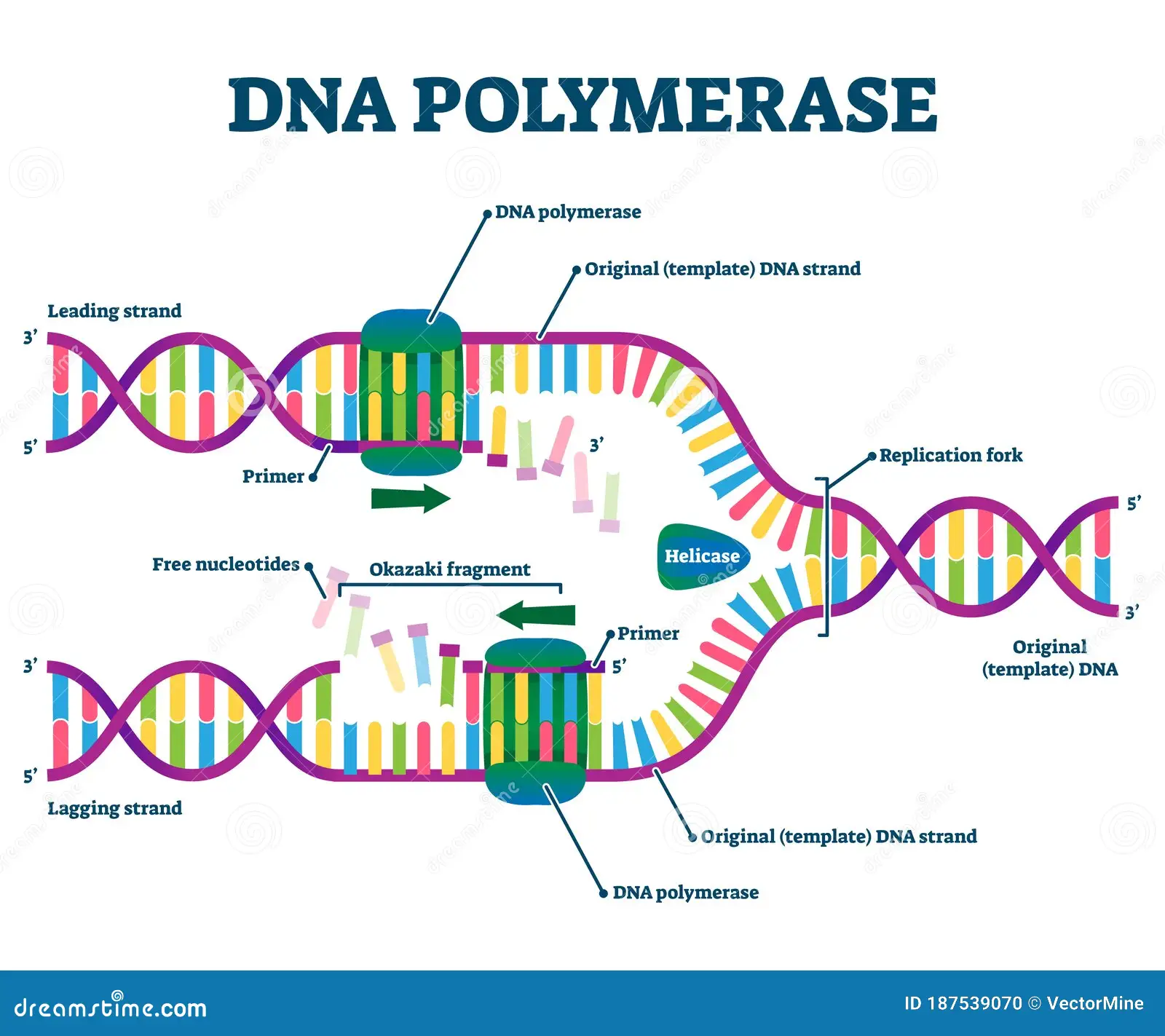
In E.coli what is the enzyme used for all new DNA synthesis?
DNA Polymerase III
except for the replacement of RNA primers
DNA I replaces the primers.
What is the function of the other DNA polymerase?
replaces the RNA primer met DNA
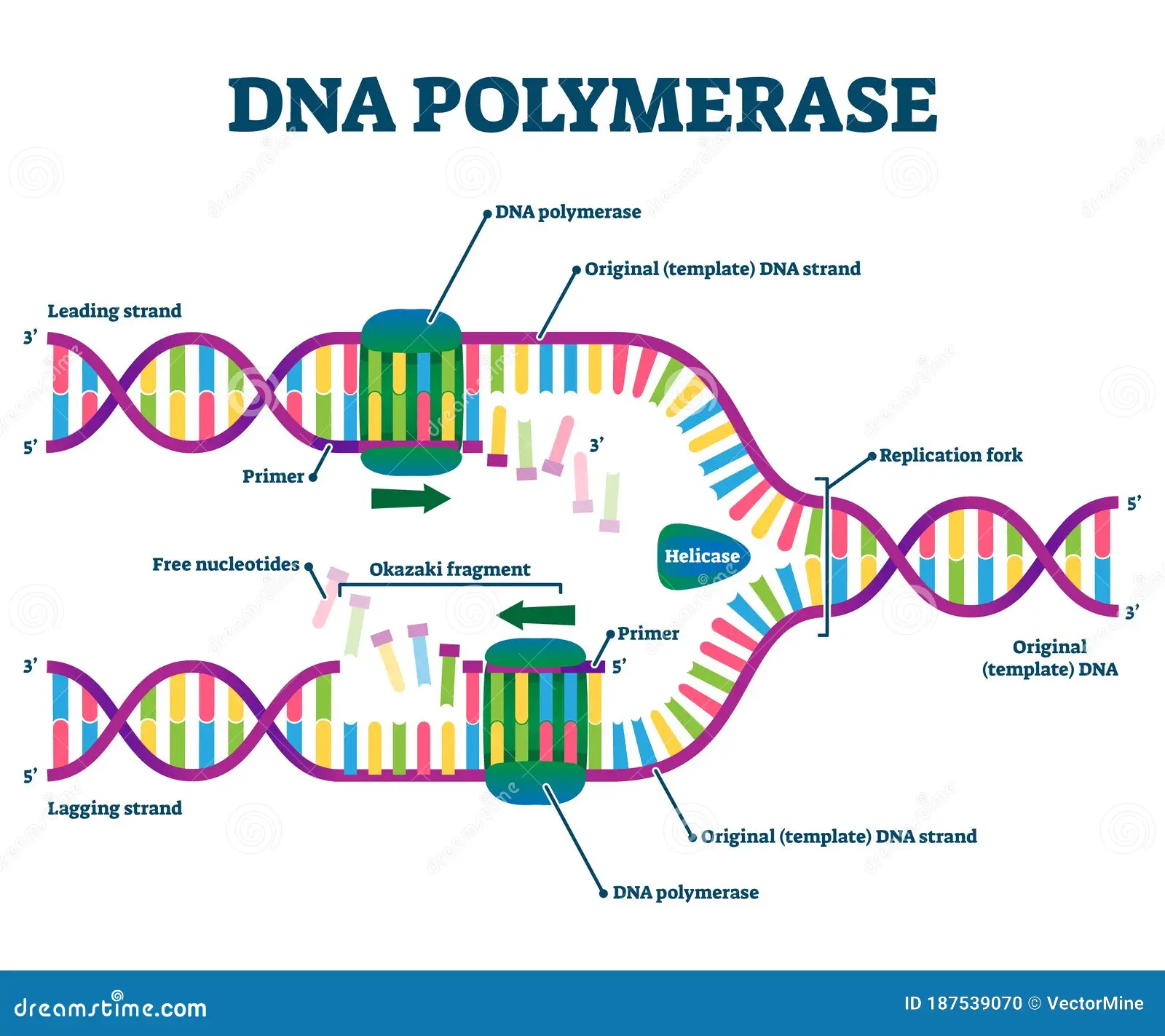
Function of DNA Ligase
seals the gap left between Okazaki fragments after the primer is removed.
The okazaki fragments join and the new lagging strands become longer and longer
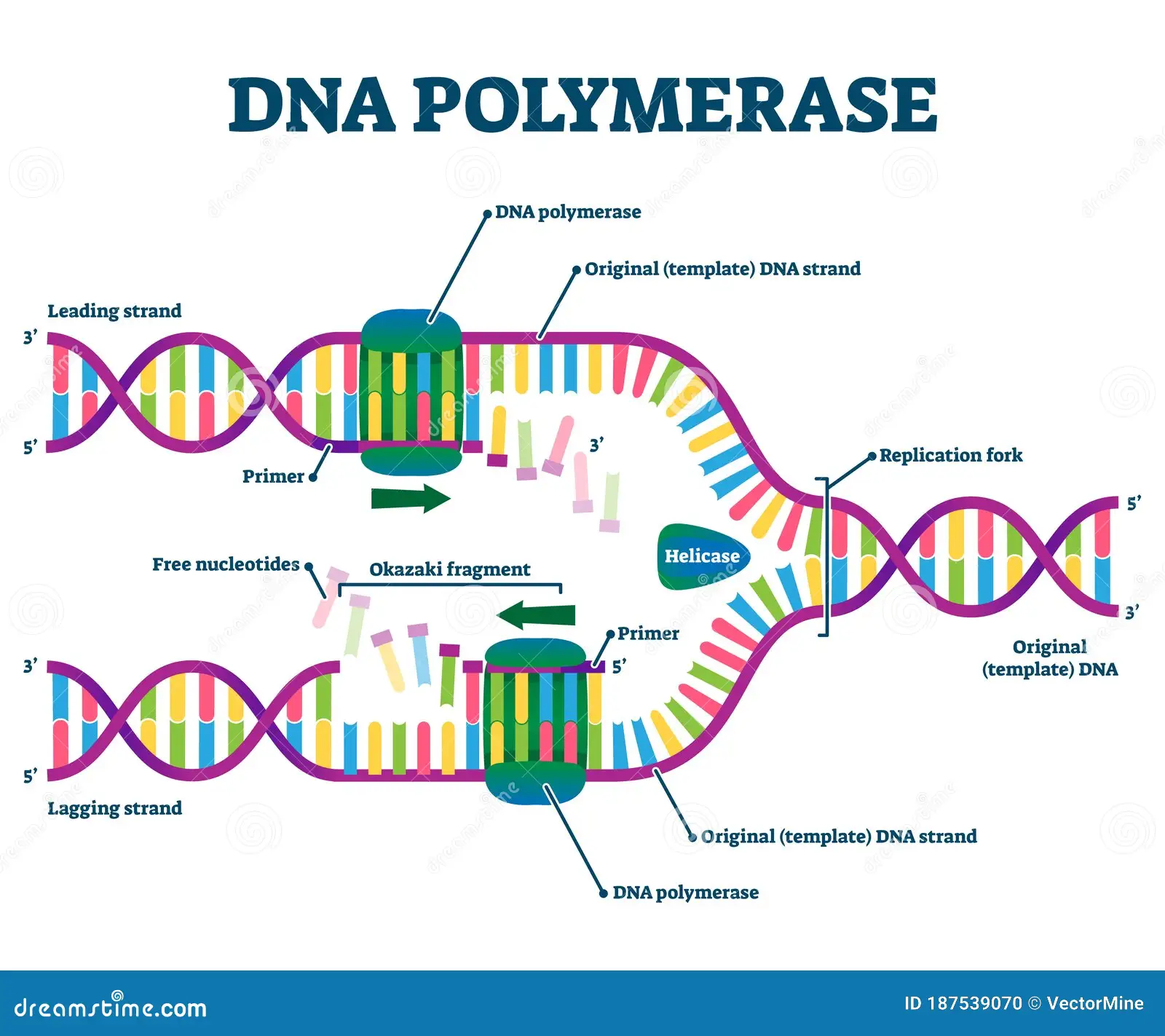
Helicase
located at the replication works; helps unwind the DNA double helix
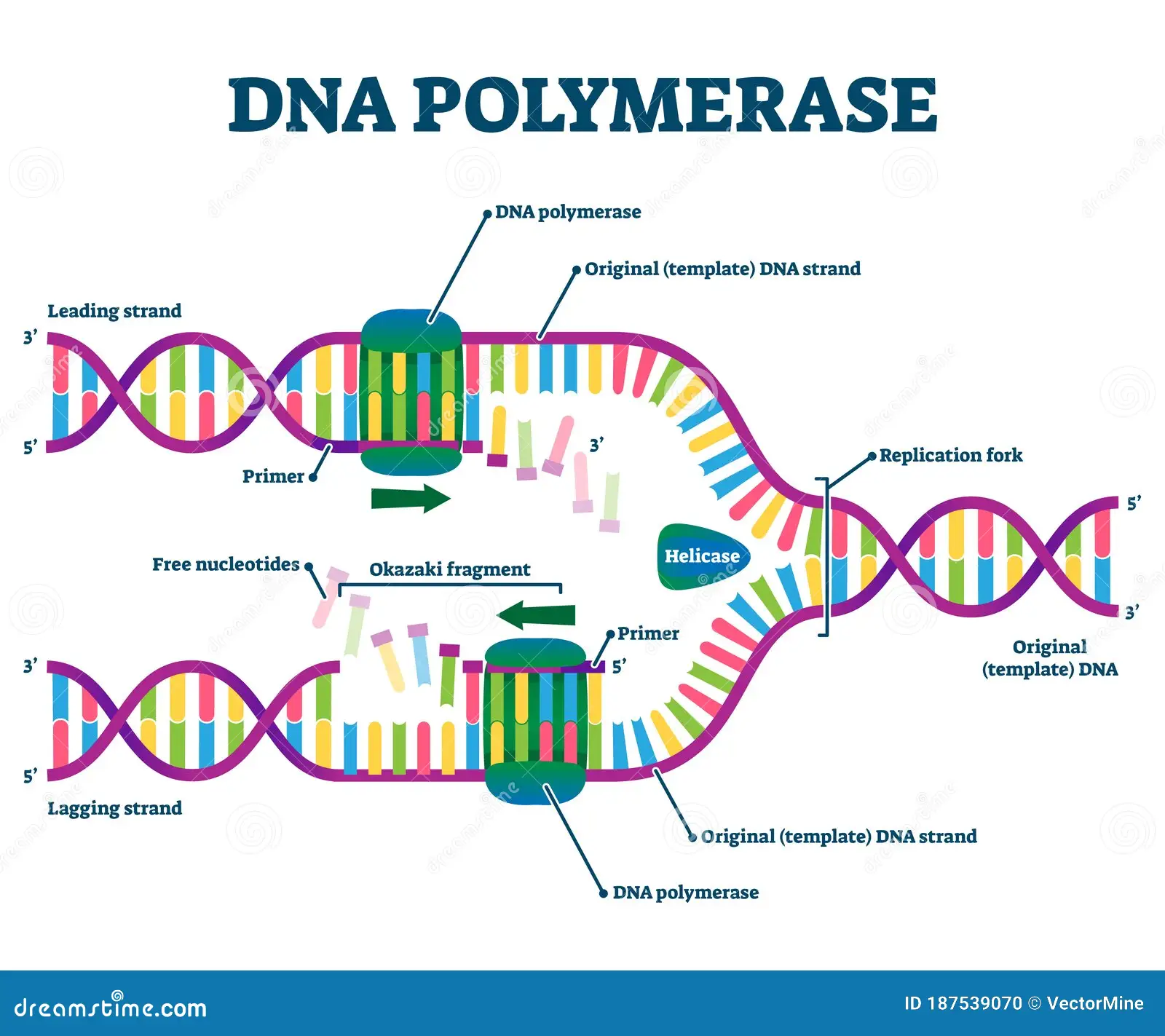
lagging strand
The new DNA stand made discontinuously in the direction opposite to the direction in which the replication fork is moving.
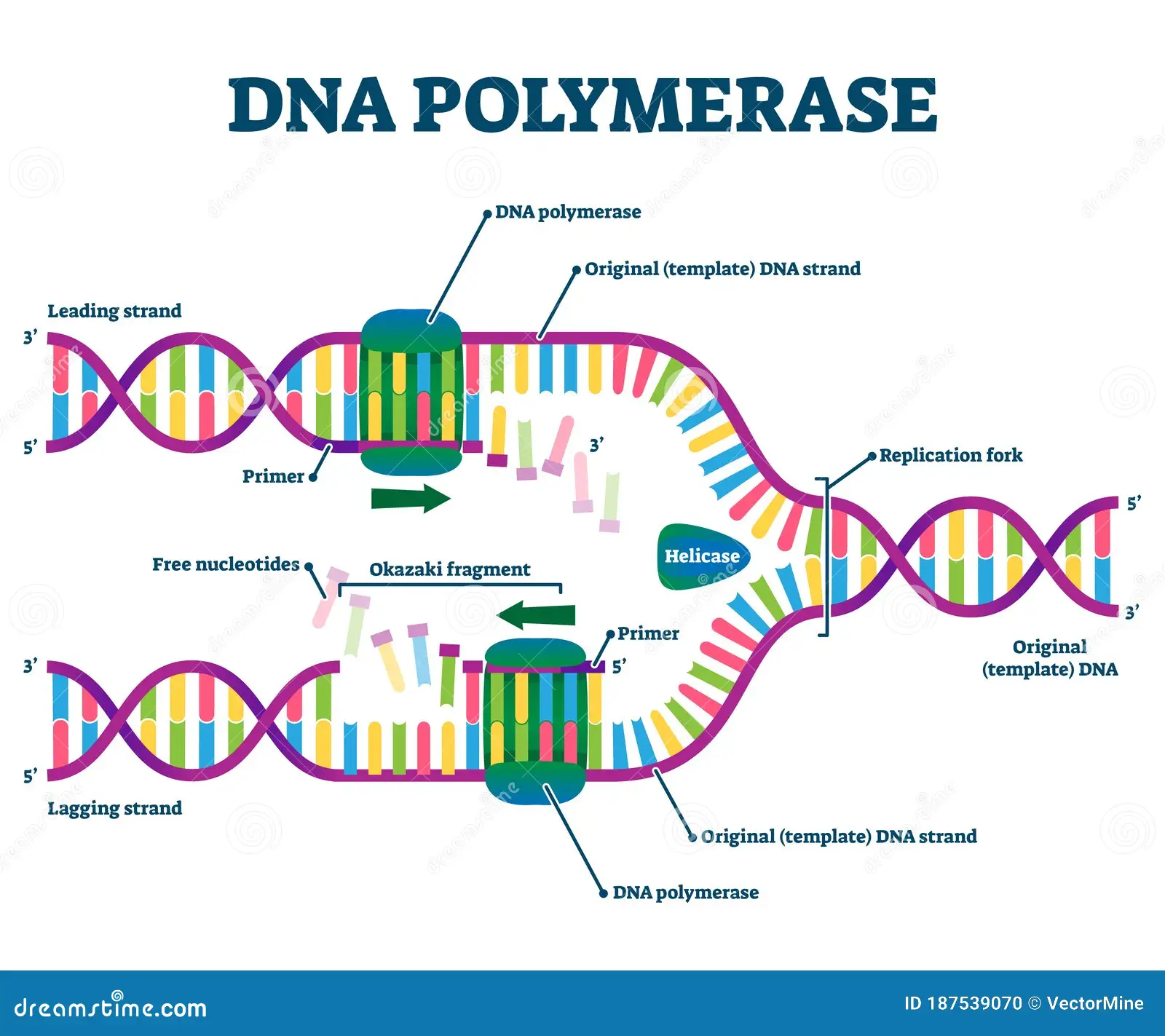
leading strand
the new DNA strand which is made continuously in the same direction as movement of the replication fork.
Moves 5’ to 3’
primase
Primase
Location: Wherever the synthesis of a new DNA fragment is to commence.
Function: DNA polymerase cannot start the synthesis of a new DNA chain, it can only
extend a nucleotide chain primer. Primase synthesizes a short RNA chain that is used as
the primer for DNA synthesis by DNA polymerase.
Whats SSB
Single strand binding protiens
• Location: On single-stranded DNA near the replication fork.
• Function: Binds to single-stranded DNA to make it stable.
Which direction does DNA replication take place
5'-end to 3'-end.
What base ratios are =1 for in a double stranded dna molecule?
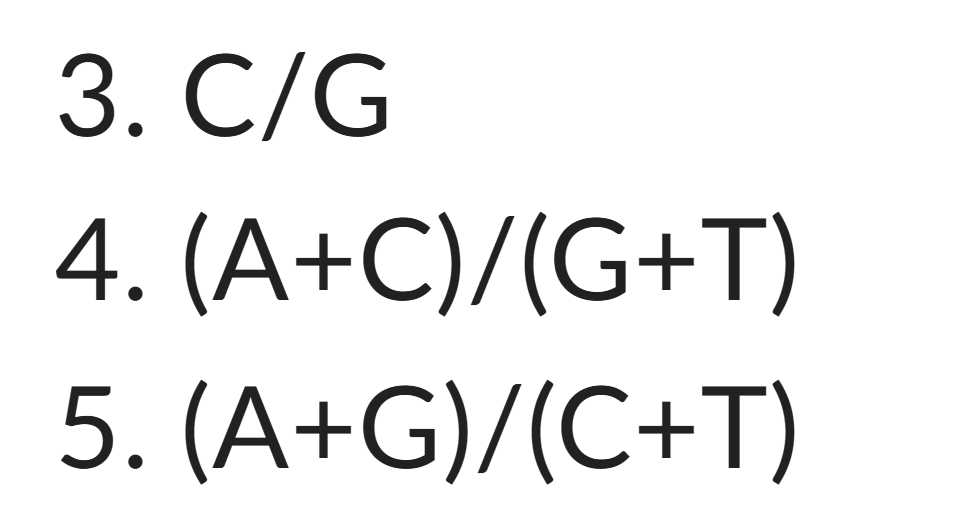
Messenger RNA
coding sequences that can be translated into proteins
small nuclear RNA’s (snRNAs);
together with several proteins they form the splicing complex
ribosomal RNA’s
together with various proteins they constitute the ribosome, where translation happens
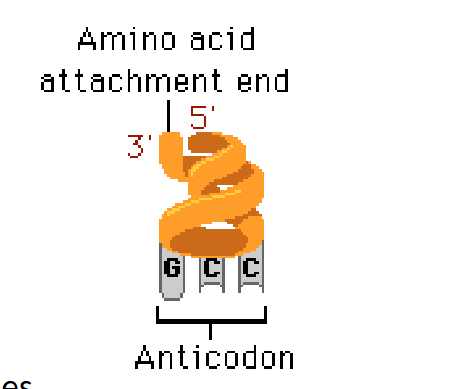
transfer RNA’s (tRNA’s);
couple, activate and transport amino acid residues to
the ribosome
• tRNAs are encoded by tRNA genes.
• All tRNA molecules are similar in size and shape.
• All tRNAs hebben CCA at the 3' end to which the amino acid attaches.
• At the other "end" of the tRNA molecule is the anticodon, which, during translation, "reads" the matching codon on the mRNA.
Which enzyme carries out transcription?
RNA polymerase
How is transcription similar to and different from DNA replication?
Both use DNA as a template, but replication copies the entire DNA and both strands, while transcription copies only part of the DNA and only one strand.
In which direction is RNA synthesized, and in which direction is DNA read?
RNA is synthesized 5’ → 3’, while DNA is read 3’ → 5’.
What determines the one-way direction of transcription?
DNA structure, RNA polymerase structure, and nucleic acid polarity.
What is the role of the sigma factor in prokaryotic transcription?
t helps RNA polymerase recognize the promoter of a gene.
What happens to mRNA in prokaryotes after transcription?
It is directly translated into protein because both DNA and mRNA are in the cytoplasm (no nucleus).
What is the first processing step of mRNA in eukaryotes?
Addition of a 5’-cap (a modified guanine nucleotide).
What is added to the 3’ end of eukaryotic mRNA?
A poly(A) tail, made of many adenines, added by poly(A)-polymerase.
What sequence signals the addition of the poly(A) tail?
The polyadenylate sequence.
What are introns and exons in mRNA?
Introns are non-coding sequences; exons are coding sequences.
What process removes introns van mRNA?
Splicing.
done through snRNAs (and associated proteins).
What’s the sigma factor?
a subunit found in rna polymerase of prokaryotes
What’s the start codon?
AUG
What is translation?
The conversion of RNA into an amino acid chain (protein).
What is a codon?
A triplet of nucleotides that codes for one amino acid residue.
several different codons can specify the same amino acid.
Where does translation start in eukaryotes?
At the AUG closest to the 5’-cap.
How is the start codon recognized in prokaryotes?
By a ribosomal RNA binding to a nucleotide sequence located before the start codon (distinguishes the first AUG).
What molecule links amino acids to codons on mRNA?
tRNA (transfer RNA).
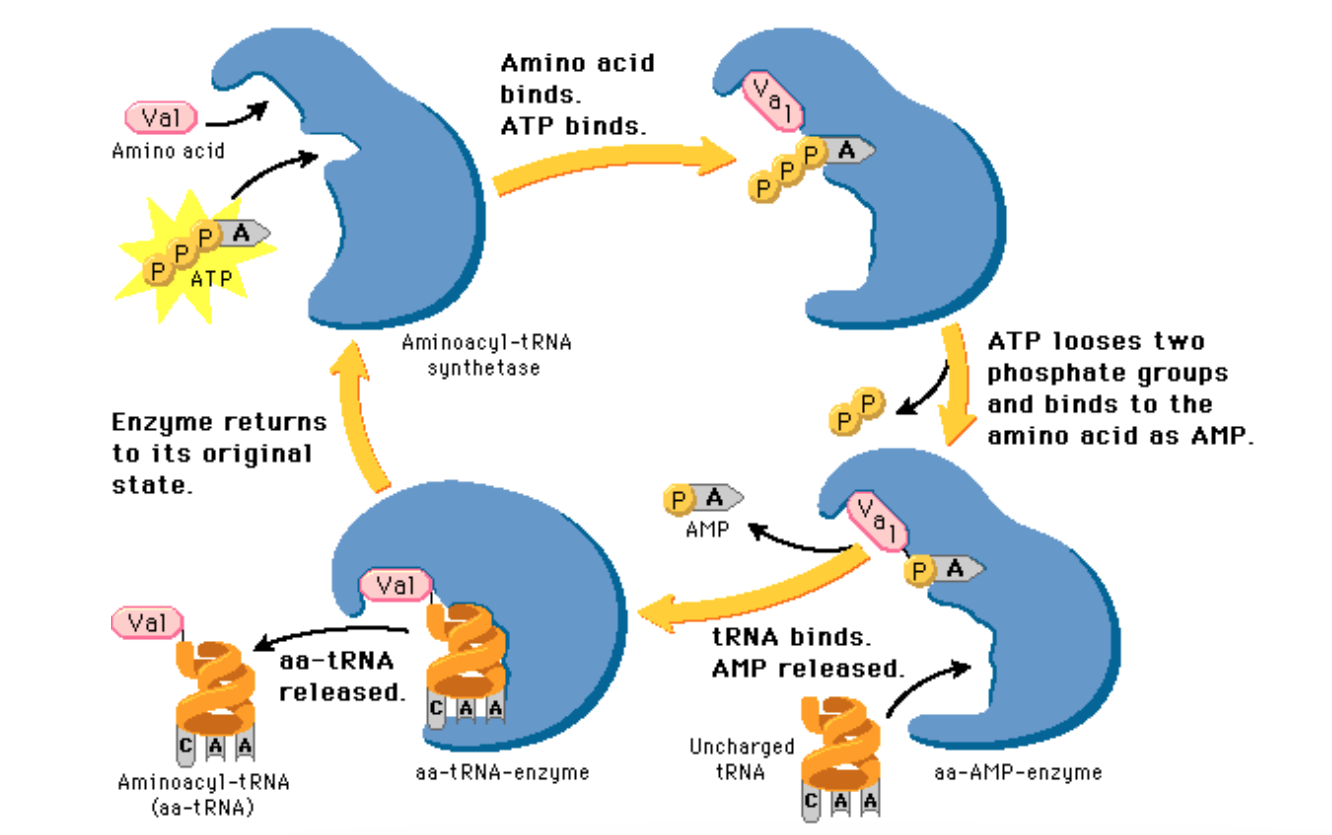
What is required to attach an amino acid to tRNA?
ATP
Which enzyme attaches amino acids to tRNA?
Aminoacyl-tRNA-synthetase.
each amino acid hebben its own aminoacyl-tRNA-synthetase.
forms a charged tRNA bound to its specific amino acid (aminoacyl-tRNA).
• All tRNAs met the same amino acid are charged by the same enzyme, even though the tRNA sequences, including anticodons, differ.
Which molecules bind to ribosomes during translation?
mRNA and tRNA.
What is a ribosome made of?
rRNA and proteins.
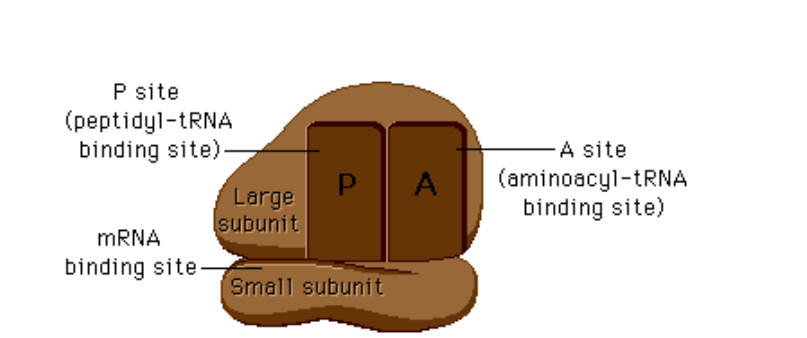
What happens at the A-site of the ribosome?
An aminoacyl-tRNA binds to the complementary codon on mRNA.
What happens at the P-site of the ribosome?
The peptide bond forms between the new amino acid and the growing polypeptide chain; tRNA carrying the chain sits here.
What happens at the E-site of the ribosome?
The uncharged (empty) tRNA exits the ribosome.
Which factor helps move tRNA and mRNA through the ribosome?
Elongation factor G (EF-G).
Where does the energy for peptide bond formation come van?
The activated amino acid attached to aminoacyl-tRNA.
How does elongation process? mention the whole process in brief
Repeated binding of aminoacyl-tRNA to the A-site, peptide bond formation, shifting of tRNA/mRNA, and release of uncharged tRNA at the E-site.
What signals the end of translation?
A stop codon (UAA, UAG, or UGA) entering the A-site.
they do not code for any amino acid.
a protein called release factor then binds to the stop codon. they Catalyzes the addition of a water molecule, breaking the bond between the polypeptide and tRNA, releasing the polypeptide.
The central Dogma
describes the two-step process": transcription and translation
by which the information in genes flows into protiens": DNA → RNA → protein
How is mRNA synthesized in the nucleus and
translated on ribosomes in the cytoplasm?
• A protein-coding gene is transcribed
into a pre-mRNA.
• Pre-mRNA is processed into a mature
mRNA.
• mRNA exits the nucleus.
• mRNA is translated on ribosomes to
produce the polypeptide chain.
What does the large subunit in an eukaryote contain?
the large subunit is 60S (named for how fast it sediments during centrifugation) and contains 28S, 5.8S, and 5S rRNAs plus about 50 ribosomal proteins.
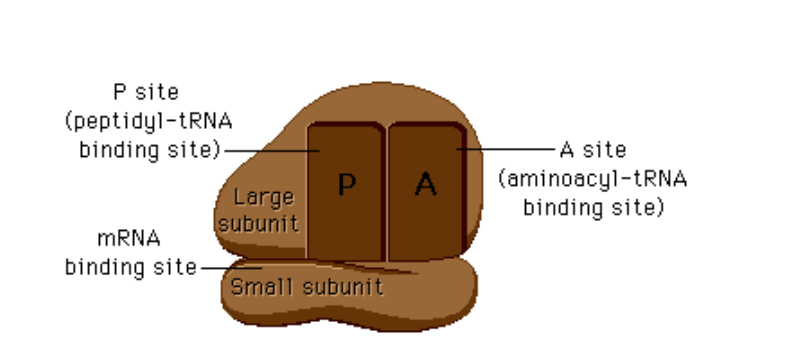
What is the small subunit in ribosomes?
The small subunit is 40S and contains 18S rRNA plus about 30 proteins.
The process of adding an amino acid to tRNA
Amino acid binds; Atp binds.
ATP looses two phosphate groups and binds to the amino acids as AMP.
tRNA binds; AMP is released
AA-tRNA is released.
Enzyme returns to its original state.
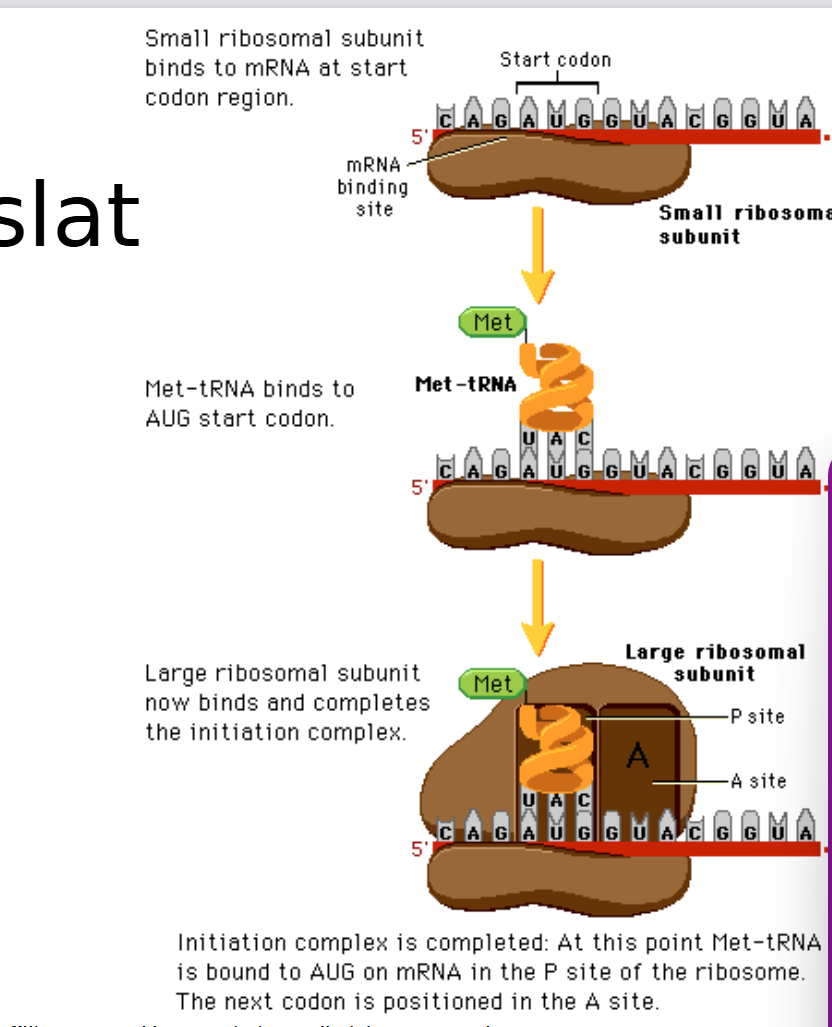
An Overview of translation; full thing
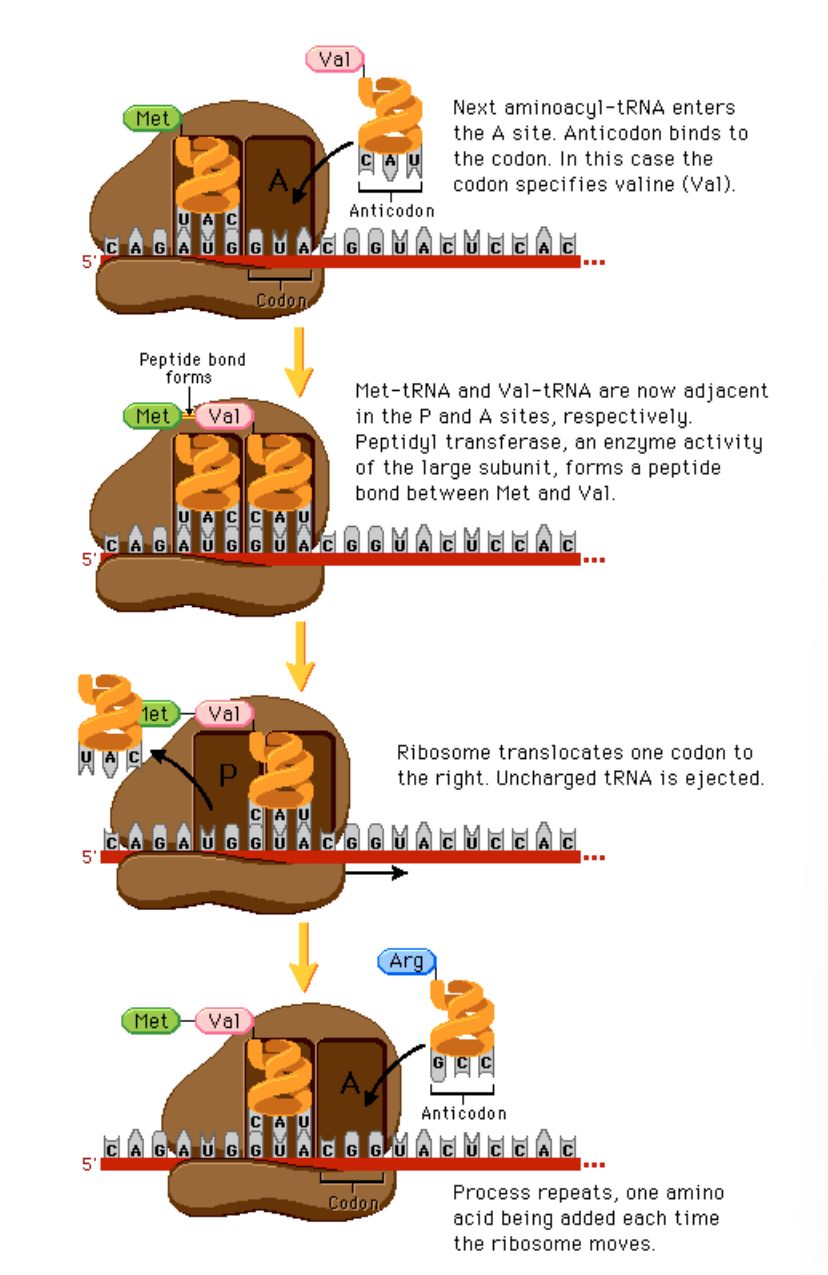
An Overview of elongation; full thing
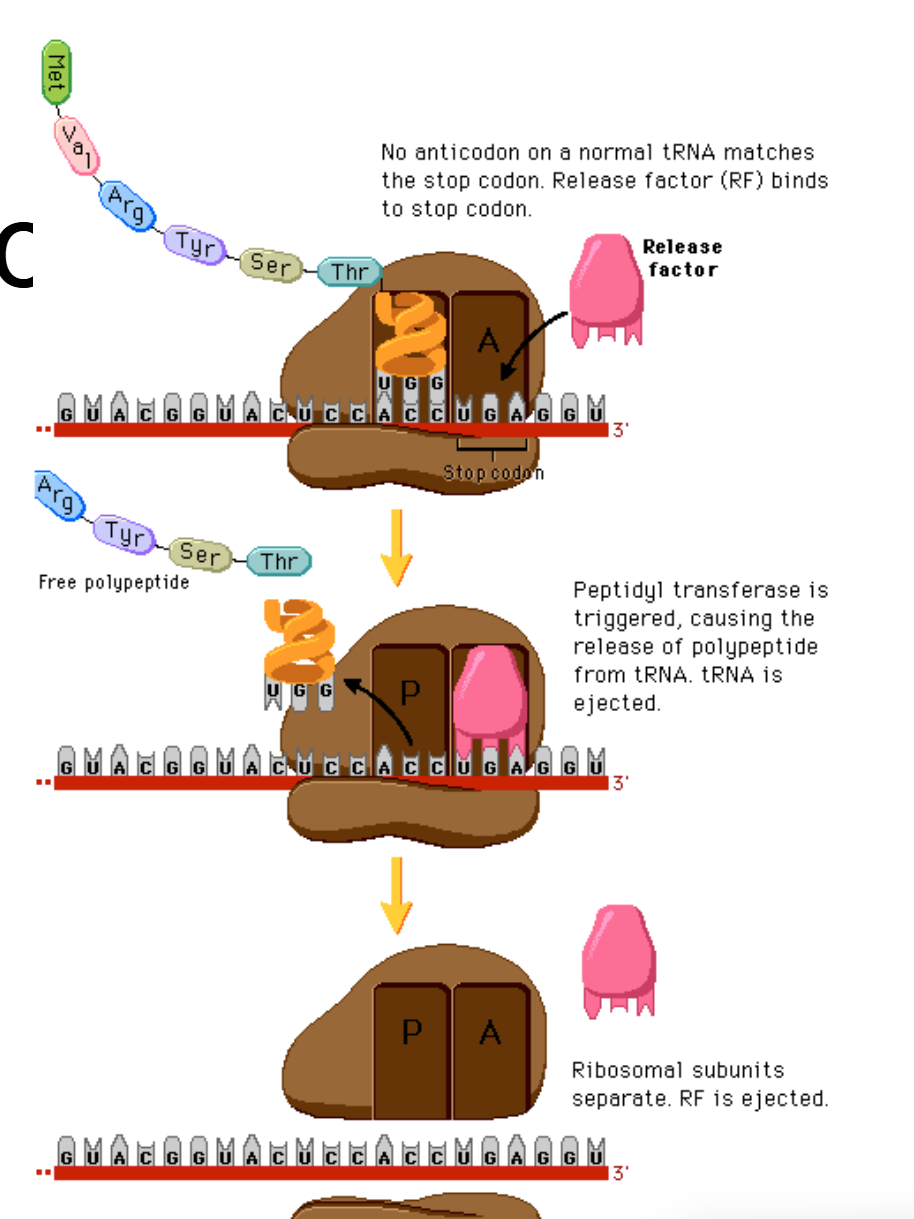
Overview of the termination of translocation; full thing
Operon
a part of DNA that consists of multiple genes, which all are under the control of the same promoter.
better definition imo:
• a cluster of bacterial genes along with an adjacent promoter that controls
the transcription of those genes.
-all genes within an operon are expressed simultaneously.
-mainly found in prokaryotes
-/upon expression they often result in formation of one mRNA molecule, which thus translates to multiple proteins.
The lac operator
A short region of DNA that lies partially within the promoter and that interacts with a regulatory protein that controls the transcription of the operon.
The lac regulator gene
Produces an mRNA that produces a Lac repressor protein, which can bind to the operator of the lac operon.
The Lac regulatory protein is a repressor because it keeps RNA polymerase from transcribing the structural genes.
Thus the Lac repressor inhibits transcription ofthe lac operon.
it negatively regulates the expression of lac genes because the operon is turned off by the regulatory protien
What is the effect of lactose on the lac operon?
When lactose is present, the lac genes are expressed because allolactose binds to the Lac repressor protein and keeps it from binding to the lac operator.
What is allolactose?
an isomer of lactose.
Small amounts of allolactose are formed when lactose enters E. coli.
it is an inducer of the lac genes.
How does allolactose function?
• The presence of lactose (and thus allolactose) determines whether or not the Lac repressor is bound to the operator.
• Allolactose binds to an allosteric site on the repressor protein causing a conformational change stopping the repressor from binding to the operator region and falls off.
RNA polymerase can then bind to the promoter and transcribe the lac genes.
What is the feedback control of the lac operon?
• When the enzymes encoded by the lac operon are produced, they break down lactose and allolactose, eventually releasing the repressor to stop additional synthesis of lacmRNA.
• Messenger RNA breaks down after a relatively short amount of time.
What is E. coli’s preferred energy source?
Glucose as It is the most available,Because enzymes for glucose metabolism are produced constantly.
When does E. coli efficiently transcribe lac genes?
Only when glucose is absent and lactose is present.
RNA polymerase efficiently transcribes the lac operon, allowing lactose metabolism.
if both are present then glucose is preffered as Lactose metabolism genes are transcribed at low levels.
Which molecules are required for the maximal transcription of the lac operon in E. coli?
Cyclic AMP (cAMP) and catabolite activator protein (CAP), which together increase RNA polymerase binding at the lac promoter.
How does glucose concentration affect cyclic AMP (cAMP) levels in E. coli? and what is cAMP derived van in E coli?
cAMP levels are inversely proportional to glucose: high glucose → low cAMP; low glucose → high cAMP.
ATP
What happens in E. coli when lactose is present and glucose is absent?
cAMP accumulates and binds to CAP. The CAP–cAMP complex binds the lac promoter and facilitates efficient transcription of the lac operon by enhancing the binding of RNA polymerase to the lac promoter.
What aspect of lac operon expression is controlled by CAP/cAMP?
The amount of lac mRNA produced.