Mol. bio ALL quiz 3
1/116
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
117 Terms
What are organelles
membrane bound compartments located within the cell
Self-replicating
Highly specialized for protein synthesis, segregation/transport, energy production, & protein degradation
Allows hydrophobic things to hide from the aqueous environment
Cytosol & mitochondria are the MAJORITY of the cell volume
Compare the rough and smooth ER
Rough ER | Smooth ER |
|
|
What is the ER
Interconnected network of tubules, vesicles, and cisternae (network)
Transportation system of eukaryotes
Protein translation
Folding & transport of proteins to be used in some cell membrane or to be exocytosed from cell
Rebuilding nuclear envelope after cytokinesis
N-linked glycosylation of proteins
Lots of proteins to help with folding
What is the Golgi apparatus (function, structure, purpose)
synthesis of carbohydrates, sorting and exporting ER products, synthesis of glycosaminoglycans (GAGs) (protein modifications)
Function: Primarily modifies proteins delivered from the rough ER through glycosylation & phosphorylation
Important in processing proteins for secretion
Involved in creating lysosomes & endosomes
Structure: Composed of membrane-bound stacks known as cisternae
Between 5-8
Has 5 functional regions (each has specific enzymes on membrane surface
Cis-golgi network
Cis-cisterna
Medial-cisterna
Trans-cisterna
Trans-Golgi network
Proteins move sequentially through the 5 regions
Purpose: adding things to protein chain to help it communicate or reach destination (processing)
Secondary oligosaccharide processing
Glycosylation = necessary for protein folding
Describe secondary oligosaccharide processing and glycosylation (in Golgi)
Secondary oligosaccharide processing = oligosaccs added in ER
Further glycosylation and phosphorylation occurs in Golgi with different enzymes at each region of Golgi (mucins and proteoglycan formation)
Glycosylation = necessary for protein folding
Resistance to proteolytic digestion, sugars can be recognized by lectins important during development and cell-cell adhesion
n-linked = can be done in golgi or ER
o-linked glycosylation = specific on Threonine oxygen
What are lyosomes
intracellular digestion
Carry out digestion function with acid hydrolases (~40)
Cell protected from lysosmal function by the lysosomal membrane
Highly glycosylated & pH environment in lysozyme, making hydrolytic enzymes non-functional
Lysosomal pH = maintenance of lysosomal pH via vacuolar H+ ATPase that pumps hydrogen ions into the lysosome
What are endosomes
Before lysosomes form, endocytosis forms early endosomes and then late endosomes and eventually lysosomes
On the way to lysosomes, endocytosed material must first pass through a series of organelles
called endosomes
What are transport vesicles
form specialized coated regions of membranes
Coats include Clathrin (P -> G), COPII (ER -> G), & COPI (G -> ER)
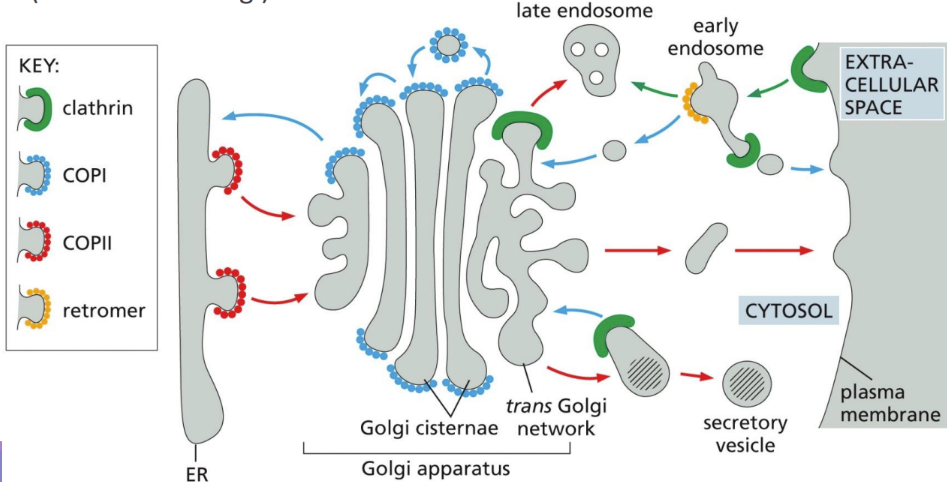
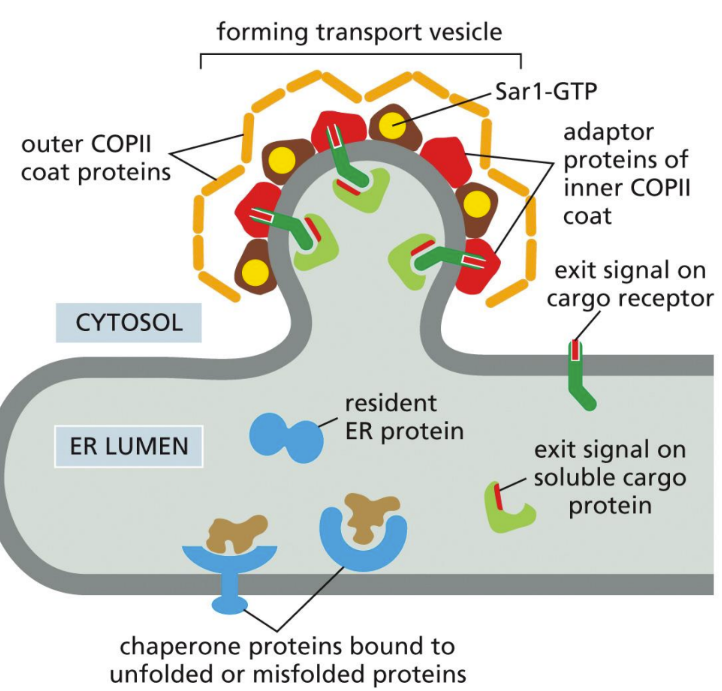
What are COPII-coated vesicles
ER to Golgi (Sar 1 -> adaptor protein -> outer coat proteins -> vesicle formed)
Active Sar 1-GTP (membrane bound to ER) recruits COPII adaptor proteins and causes the membrane to deform
The adaptor proteins will then recruit the outer coat proteins to help form the bud
Exit signal on cytosolic tails of cargo proteins interact with membrane cargo receptors and soluble ER proteins & complex into budding transport (interacting with Sar 1-GTP and COPII proteins)
A subsequent membrane fusion event pinches off the coated vesicle
Vesicles shed COPII coat and fuse with each other with SNARES, forming vesicular tubular clusters
What are COPI-coated vesicles
Proteins without this signal can leak out of the ER
Retrieved by COPI proteins with retrieval signals (KDEL & KKXX)
KDEL receptors in tubular clusters and Golgi capture the escaped protein and carry them back to ER with COPI coated vesicles
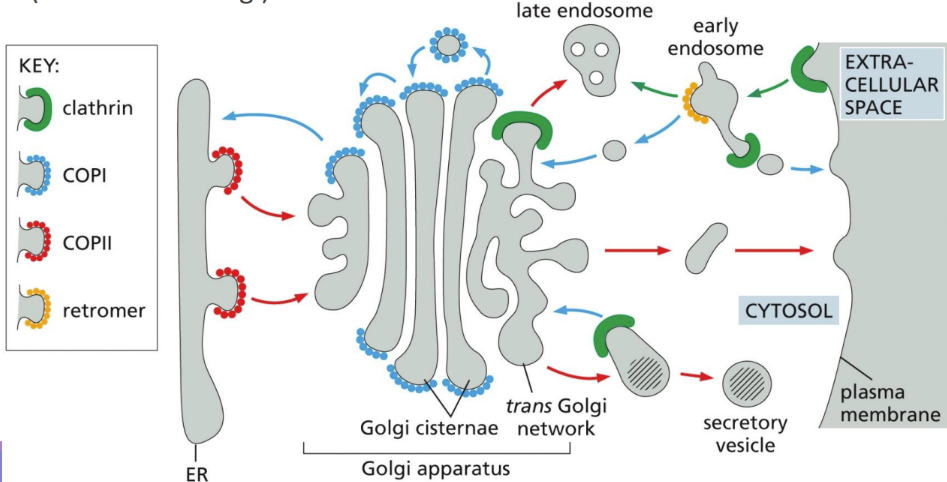
How does Clathrin work
Golgi to the plasma membrane
Receptors on trans-Golgi for M6P put clathrin-coated protein and transport it to the lysosome
How does ER to Golgi work
If proteins are properly folded & assembled, they’ll be packaged into COPII coated transport vesicles that bud from the ER membrane and go to the Golgi
Proteins get into the transporter by...
Selective process = exit (transport) signal on proteins that COPII recognizes
Proteins without this signal can leak out of the ER
Retrieved by COPI proteins with retrieval signals (KDEL)
KDEL receptors in tubular clusters and Golgi capture the escaped protein and carry them back to ER with COPI coated vesicles
Loss of transporters can lead to various disorders (fat absorption, protein secretion)
Some proteins only need a receptor on the ER membrane (because are soluble)
How does the process to the golgi work? What is homotypic fusion?
After the vesicle forms from the ER, the COPII proteins are released and the vesicles begin to fuse forming vesicular tubular clusters
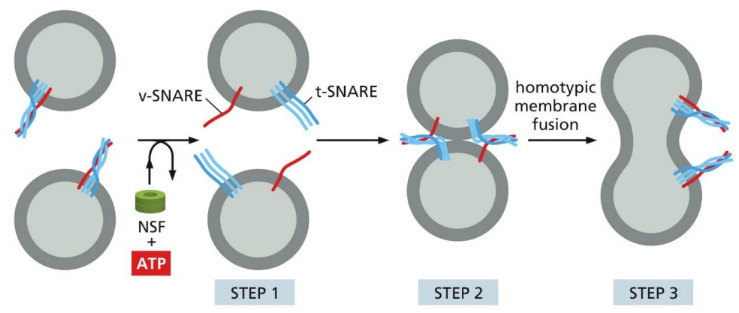
Homotypic fusion = utilizes SNARE proteins (a t-SNARE and v-SNARE are required to force lipid bilayers closed and expel water)
How does homotypic fusion work
Homotypic fusion = utilizes SNARE proteins (a t-SNARE and v-SNARE are required to force lipid bilayers closed and expel water)
SNARE = transmembrane proteins that catalyze membrane fusion reactions
v & t- SNARES are wrapped together
NSF protein (N-ethylmaleimide sensitive factor/fusion protein) prys v and t-SNARE apart
v & t-SNARE bind with other v and t-snare on a different vesicle
They fuse/wrap together
Repeated
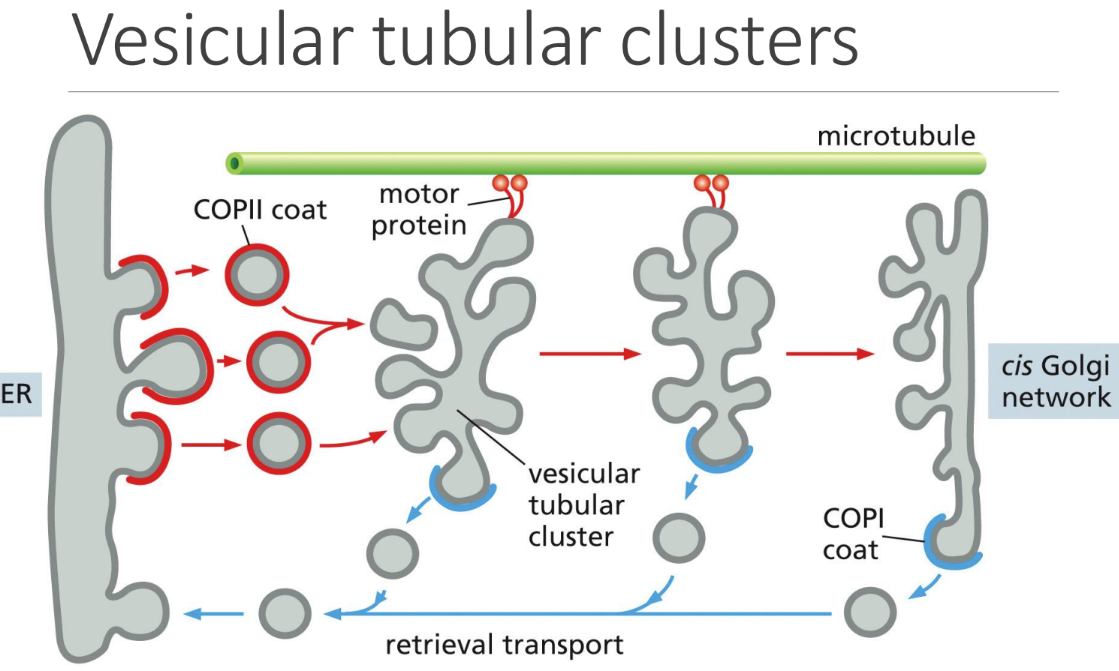
What are vesicular tubular clusters
Vesicular tubular clusters = multiple vesicles fuse together so only 1 large one is transported to the Golgi
Pulled along microtubules to Golgi Cis face
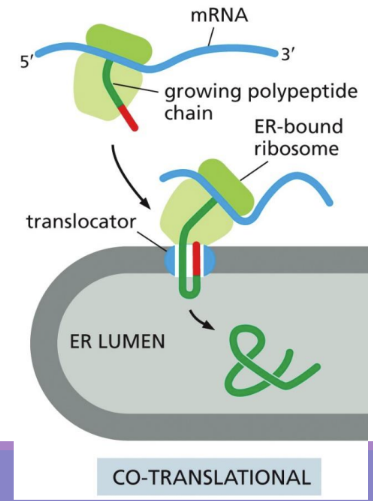
What is Co-translation translocation
Co-translational translocation = in rER, proteins are imported before they are completely translated
Ribosome is attached to ER (via special particles and sequences)
uses SRP
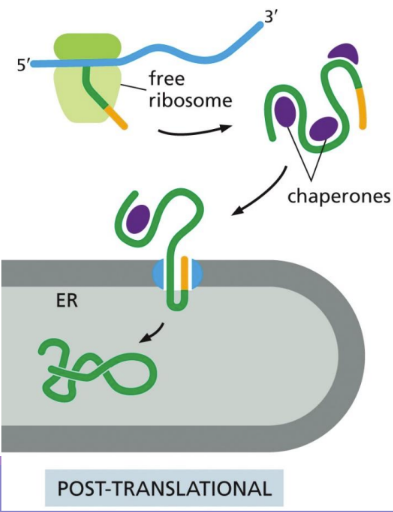
What is post-translational translocation
Post-translational translocation = Free ribosomes complete protein synthesis and release prior to the protein being translocated to a mitochondrial/nuclear membrane (NOT ER PROCESS)
What is a signal sequence
a short sequence of amino acids at the N-terminal that direct secreted proteins to the ER & is then cleaved off by signal peptidase
Different for different polypeptides but contains several nonpolar AAs
Found in both transmembrane (destined for ER) and water-soluble (secreted or stay within organelle) proteins
Used in both co-translational and post-translational translocation
Not part of the final protein because it gets cleaved off by signal peptidase
Signal peptidase cleaves of N-terminus
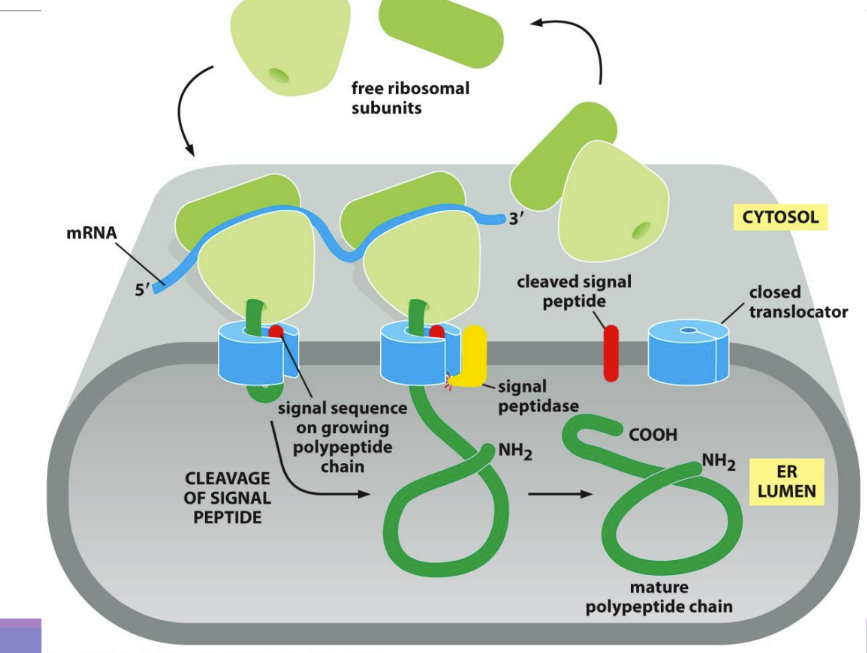
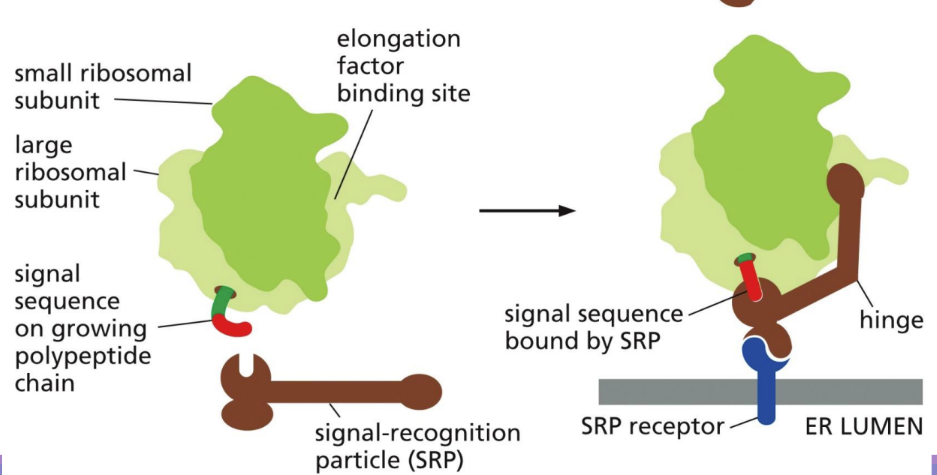
What is a signal recognition particle (SRP)
guides signal sequence to ER translocator and SRP receptor (integral membrane protein on ER membrane – in blue)
Has a signal sequence binding site lined with methionine (flexible) allowing it to accommodate many signal sequences
One end binds to signal sequence & other blocks E site (stopping translation)
ONLY USED IN CO-TRANSCRIPTIONAL TRANSLOCATION
Contains an RNA and 6 protein subunits
Rod-like shape that wraps around the large ribosomal subunit
Where do different parts of membrane proteins remain
Membrane proteins:
Alpha helicies = Hydrophobic segments in the peptide chain are anchored in the protein membrane by a stop-transfer signal (stay inside away from the aqueous environment)
Water soluble proteins = stay in the cytosol
Single-pass transmembrane proteins = pass through the membrane only once (hydrophobic segment stays in the membrane)
How does the SRP work
SRP (with ribosome/signal sequence) binds to SRP receptor on ER membrane
The ribosome (with the polypeptide chain) moves and binds to the translocator on the ER membrane
Peptide is completed within ER cistern
*ONLY WITH CO-TT
What happens after the ER
Many proteins stay in the ER
Have a KDEL (4 amino acid sequence = retention signal) sequence at C-terminus
Also serves as a retrieval signal if accidently gets sent out of ER
Must be folded properly to stay in the ER
Ex: BiP = hold proteins for degradation in the ER
Calnexin & calreticulin (heat shock proteins)
Others leave the ER (to the Golgi for further processing)
Processed by adding oligosaccharides (glycosylation)
What is glycosylation
adding oligosaccharides
Occurs at the amine group of an Asparagine
N-linked
Results in the removal of a mannose and glucoses (“trimming”)
Important for transmembrane proteins on extracellular side for cell-cell interactions
Oligosaccharide tags onto 90% of glycoproteins & are markers for protein folding
How does movement through the golgi work
Moving through the Golgi:
Unclear process but it is known that vesicular transport forms into what is needed to get the job done
Directional
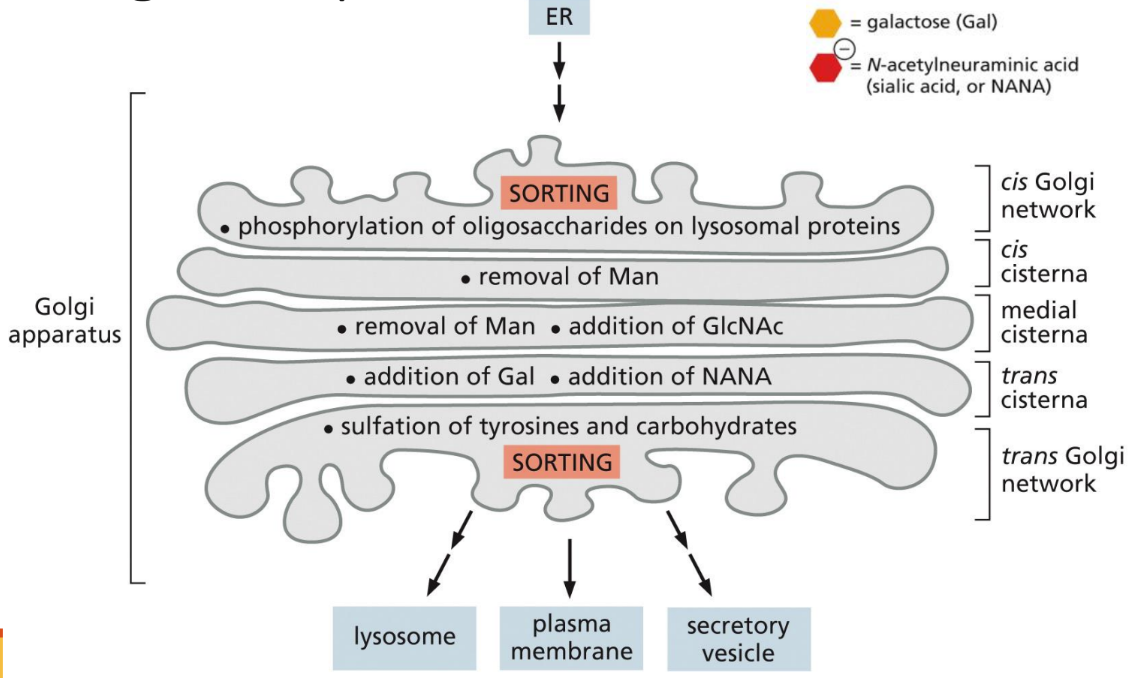
What are secretory vesicles
Secretory vesicles bud from trans-Golgi network packaged into secretory vesicles
Secretory proteins first aggregate in vesicles
Vesicle contents released in response to certain signals
What is proteoglycan assembly (and proteoglycan structure)
adding sugar groups to proteins (for stability/rigidity)
Golgi proteoglycans assembled in the Golgi
For protection, communication, and adhesion
Glycosyl transferase enzymes link oligos to hydroxyl groups of certain amino acids in a protein backbone (addition of GAG chains)
Proteoglycans are important in extracellular matrix
Proteoglycan structure:
Hyaluronic acid backbone
Link proteins connect core proteins
O & N-linked oligosaccharides are branching off
What is Charcot-Marie-Tooth disease
motor nerve disorder appearing first in the legs and then arms. Eventually have sensory symptoms (can’t feel in limbs)
Loss of peripheral sensory and motor nerves (myelin disorder)
Damaged myelin sheath
Common cause = duplication region in chr. 17p (has gene that makes myelin)
40 different possible genes involved
What is Multiple sclerosis (MS)
CNS demyelinating disease (autoimmune)
No nerve impulses because myelin affected
Cause is unknown, but ALL possible causes stimulate the ERSR in myelin producing cells
Virus? Gene defect? Both? Cells trying to remyelinate axons?
What is Achondrogenesis
congenital disorder that results in defects in the structure of the golgi apparatus
Mutation in TRIP11 gene
Affects cartilage and skeletal development
Affected infants have extremely short limbs, narrow chest, short ribs that fracture easily, & lack of normal bond formation in skull/spine/pelvis
What is DEAF 1555
hearing loss dur to an A to G mutation in mitochondrial DNA
What is MELAS 3242
deafness, stroke, or diabetes
What is LHON
optic neuropathy (blindness)
What is MERRF
myotonic epilepsy and ragged-red fiber disease
What is NARP 8993
Leigh’s encephalopathy
What is ADPD 4336
Alzheimer/Parkinsons
What do problems in the ER lead too
too many mis/unfolded proteins accumulating in/near ER causing it to become stressed
Can start an apoptotic response within cells to eliminate affected cells
ERSR or UPR = inhibits translation of many proteins, but upregulates chaperones (issue with BiP)
Occurs mostly in nervous system where myelinating cells produce much plasma membrane in the myelinating process
Involved in disorders like CMTD & MS
What do disorders in the Golgi lead to
Inclusion-cell disease = Group of congenital glycosylation disorders caused by mutation in genes encoding glycosylating enzymes or transport proteins (lysosomal storage disease because no hydrolases in lysosomes)
Lethal by age 2
Proteins are excreted instead of going to lysosomes
Alzheimer disease = neuronal golgi fragments and atrophies (amyloid build)
What do mitochondrial diseases lead to
blindness, heart failure, GI problems, tremors, poor balance
Not enough energy produced
Cells and organs don’t function
Enough mitochondria must be affected for symptoms to show up (more than just 1 or a few)
Tissues highly dependent on oxidative respiration are the most affected
Brain, heart, muscle
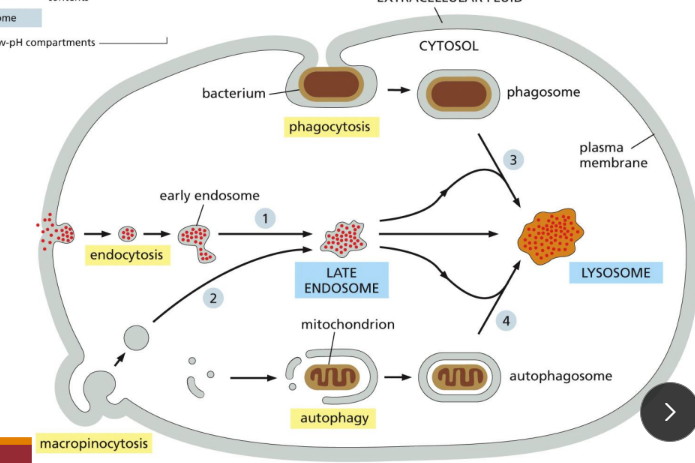
What are the lysosome degradation pathways
Endocytosis = cells take in components of the plasma membrane and extracellular space and deliver them to internal compartments called endosomes
Phagocytosis = the plasma membrane is directed to wrap around the particle to be engulfed until it fuses with itself, resulting in an enclosed phagosome inside the cell (bacteria)
Autophagy = This pathway engulfs parts of the cytosol or whole organelles into a newly assembled compartment, which then fuses with lysosomes to deliver its contents for degradation
Deletion of obsolete cell parts via lysosomal digestion
Macropinocytosis = a process whereby the plasma membrane protrudes from the cell and engulfs a portion of the surrounding extracellular fluid into a macropinosome.
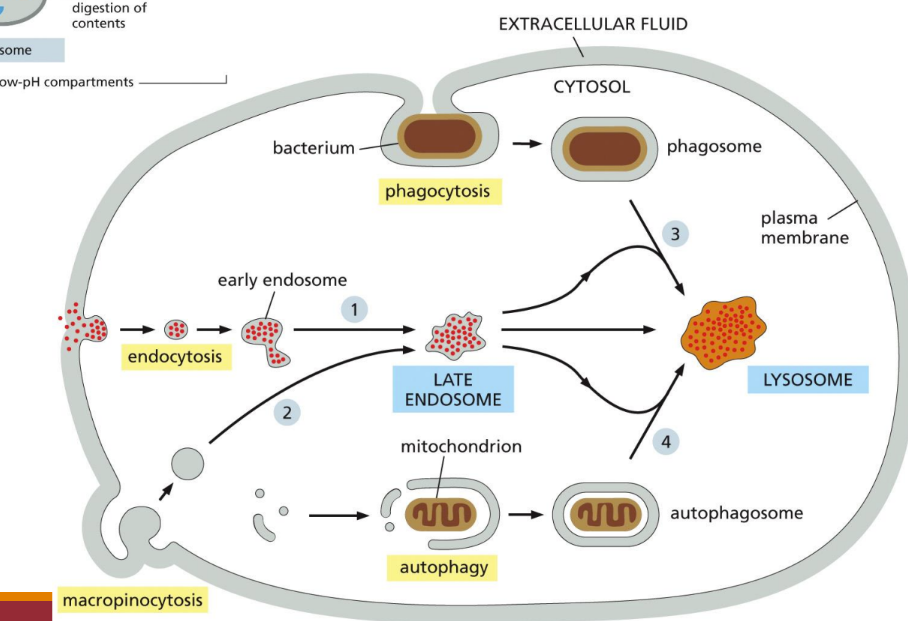
What is lysosomal hydrolase sorting
Enzymes have a mannose-6-phosphate group attached in N-linked oligosaccharides
Receptors on trans-Golgi for M6P put clathrin-coated protein and transport it to the lysosome
What is the mitochondria
organelles that generate most of the cell’s supply of ATP
Free radical generation, apoptosis control, cell growth, & cell cycle regulation
~2,000 per cell
Composed of compartments that carry out specialized functions (multi-layered)
Outer membrane, intermembrane space, inner membrane, cristae, and matrix
Involved in formation of hydrogen gradient using fat and sugar and producing water and CO2
Electron transport process done by the citric acid cycle
CAC makes 2 CO2, 3 NADH, 1 GTP, and 1 FADH2
Maternally inherited
That's why there is a different range of disease severity when mitochondria affected
What is the outer membrane of the mitochondria
contains complexes of integral membrane proteins that form channels through which a variety of molecules and ions move in and out (simple receptors)
What is the inner membrane of the mitochondria
contains 5 complexes of integral membrane proteins
NADH dehydrogenase (complex I)
Succinate dehydrogenase (Complex II)
Cytochrome c reductase (complex III)
Cytochrome c oxidase (complex IV)
ATP synthase
GOAL OF COMPLEX IS TO CREATE A PROTON GRADIENT and ultimately ATP
What is the mitochondrial matrix
contains mixture of soluble enzymes that catalyze the respiration of pyruvic acid and other small organic molecules (mtRNA (majority protein coding) & tRNA)
Pyruvic acid is oxidized and decarboxylated to produce CO2 and a fragment of acetate which is donated to oxaloacetate to form citric acid
What is mitochondrial DNA
made of 37 genes that encode for... (majority protein-coding genes)
2 ribosomal RNAs, 22 tRNAs, 13 polypeptides embedded in the inner membrane, 7 subunits that make up the NADH dehydrogenase, 3 subunits of cytochrome c oxidase, 2 subunits fo rATP synthase, & cytochrome b
What is exocytosis and the 2 types
proteins destined for cell export are in trans golgi transport vesicles that fuse with the plasma membrane of the cell
Constitutive secretory pathway
Regulated secretory pathway
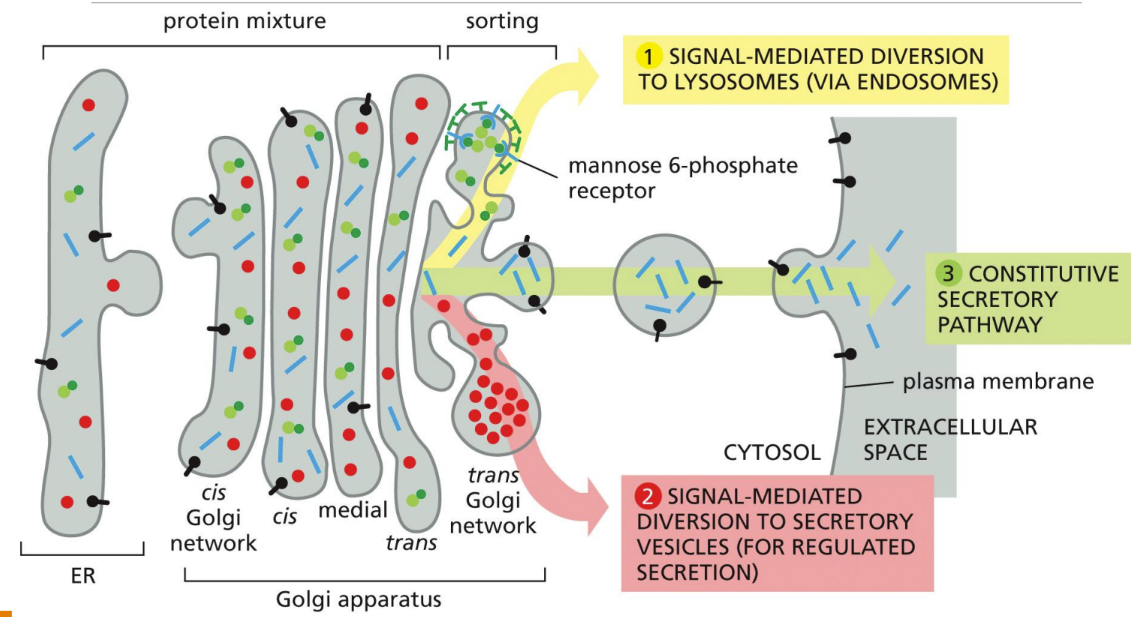
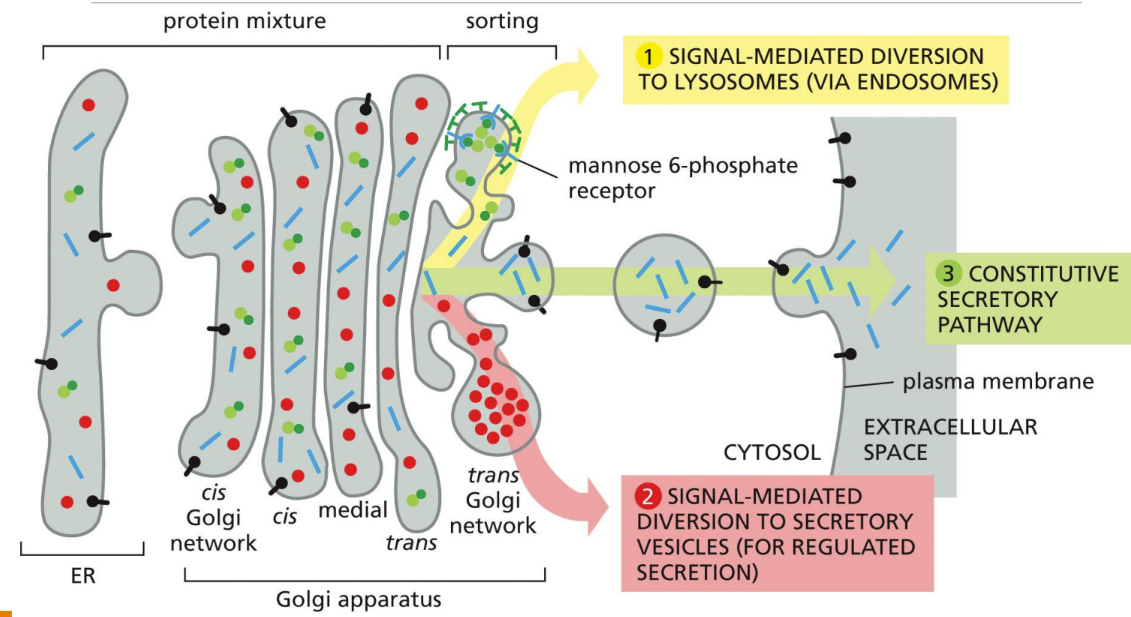
What is the constitutive secretory pathway
Constitutive secretory pathway = operates continuously in unpolarized cells (fibroblasts, WBCs)
No signals required for proteins to leave
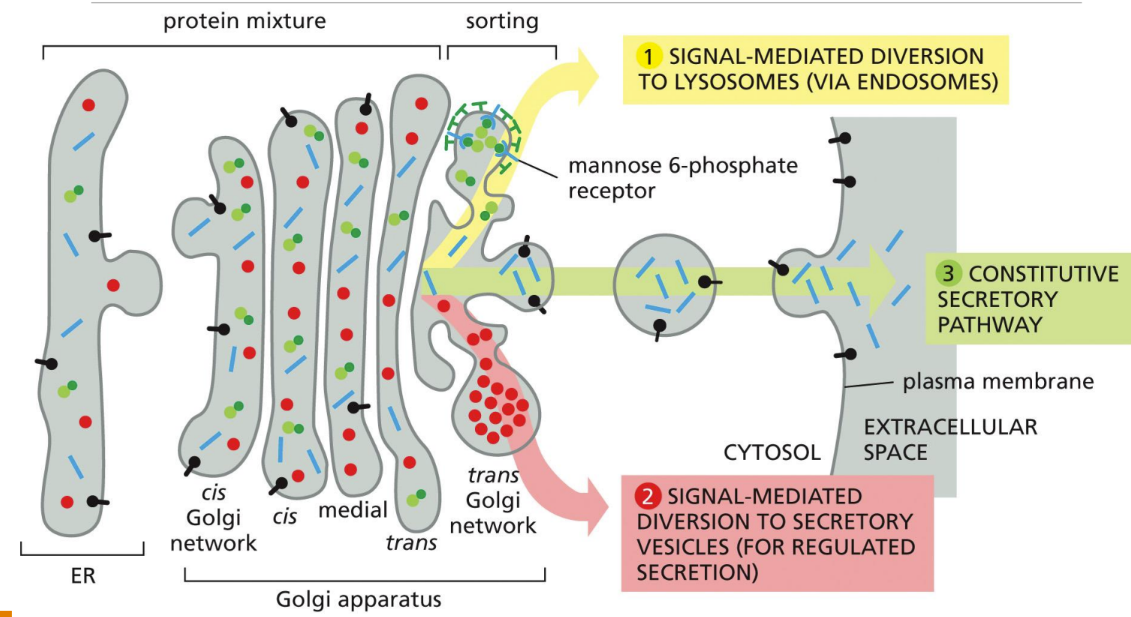
What is the regulated secretory pathway
Regulated secretory pathway = cells that secrete proteins rapidly to different domains of the cell surface (more complex) (secretion triggered)
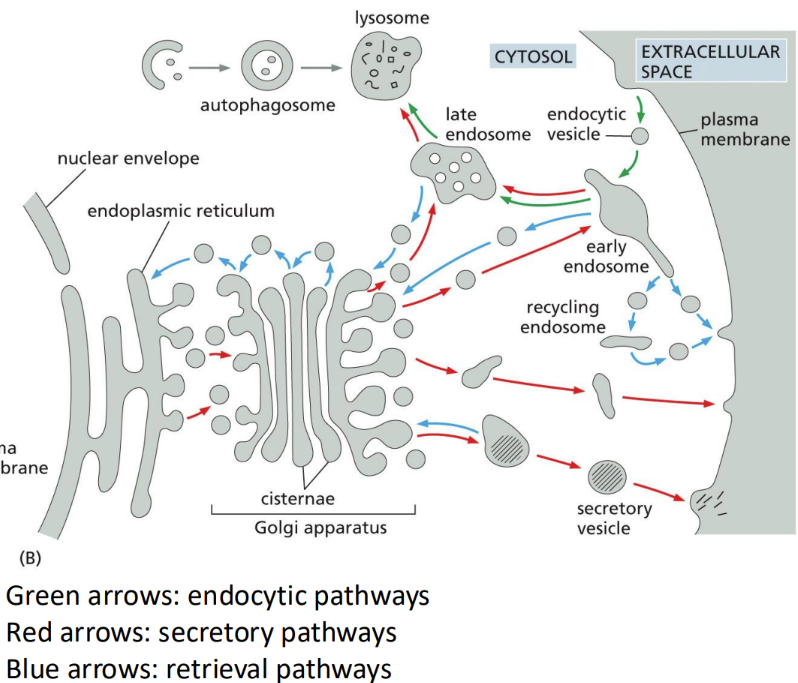
Describe the image
word
What makes up a polypeptide
Backbone = the regular structure that forms when amino acids are linked together via a peptide bond. Consists of repeating units of NHs, CHs, C=O, NH, CH etc.
Amino acid side = attached to repetitive backbone
Some are polar, nonpolar, or hydrophobic
PROTEIN FOLDING BASED ON THE DISTRIBUTION OF SIDE CHAINS AND BONDS FORMED BETWEEN DIFFERENT PARTS OF POLYPEPTIDE
There's interactions between side chains: Hydrogen bonds, Van der Waals, & electrostatic attractions
What are the hydro. interactions within a polypeptide chain/protein
Hydrophobic interactions inside
Non-polar side chains inside in the core
Hydrophilic interactions outside
Polar side chains gather on the outside
What is a primary structure
sequence of a chain of amino acids (linear)
Involves amino acid sequence
What is a secondary structure
amino acids interact with each other to fold into a repeating pattern (protein folding)
Two regular folding patterns caused by hydrogen bonding are found in parts of proteins (secondary structures)
made up of alpha helix and beta sheets
Protein domains = small units of protein that fold independently of each other
Different protein domains of a single protein perform different functions
Ex: SRC = protein kinase that adds phosphate groups to other proteins using ATP as the substrate in cell signaling (has 3 domains: 1 catalyzes & 2 are regulatory)
Polar side chains gather on the outside
Non-polar side chains gather at the core
What is an alpha helix
specific folding patterns because of hydrogen bond formation
A single helix resulting when a single polypeptide chain twists around itself to form a cylinder
Ex: Keratin (fingernail protein)
What is a beta sheet
antiparallel chains
Different amino acids join to form this type of protein structure, and it is the specific amino acids that make this structure
Ex: fibroin (silk component)
What is a tertiary structure
3D folding pattern; attractions between different secondary structures
Alpha & beta sheets connecting together
The combination of different primary and secondary structures
What is a quaternary structure
Other peptide chains interact with main protein chain
NOT all proteins reach this point
Different unconnected proteins join together to form one protein
Ex: hemoglobin made of different globins
What are fibrous proteins
filamentous proteins (considered to be intermediate filaments)
Hold things together (structural things)
Long fiber
Ex: Fibrinogen, collagen, troponin
What are globular proteins
tend to be rounded and bulky
Large, bulky & compact
Ex: hemoglobin, enzymes, plasma proteins
Why is proteins binding to ligands important
Structure = function
Biological properties are determined by what a protein binds to (physical interactions)
All proteins bind with high specificity
Ligand = substance bound by a protein (binds to receptor)
Weak, non-covalent bonds hold a ligand to a protein
Ligand-binding site = cavity in the protein surface formed by amino acids
Allows binding to be specific
What is an enzyme
proteins that bind one or more ligands (substrates) and convert them into products
Catalysts permit cells to make and break covalent bonds (speed up reactions
Ex: alpha amylase binding starches and sugars to make monosaccharides
Usually end in –ase
Types: Hydrolase, nuclease, proteases, synthases, ligases, kinases, phosphatases
What do hydrolases do
Hydrolases = catalyze a hydrolytic cleavage reaction
Types
Nucleases = break down nucleic acids by hydrolyzing bonds between nucleotides
Proteases = break down proteins by hydrolyzing bonds between amino acids
What does synthases do
synthesize molecules in anabolic reactions by condensing two smaller molecules together
What does ligase do
join together 2 molecules in an energy dependent process
What does kinase do
catalyze the addition of a phosphate group to a molecule
What does phosphatase do
catalyze the hydrolytic removal of a phosphate group from a molecule
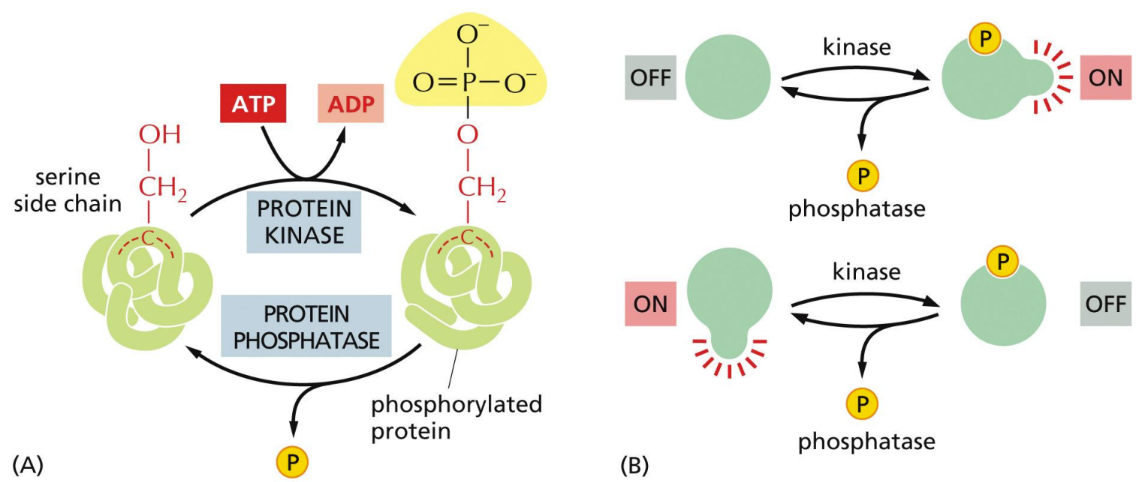
What does protein phosphorylation do
proteins regulated by adding or removing phosphate (activate/repress protein)
Adding a phosphate group causes conformational change because the charges on the protein are changed
Forms binding sites for other molecules
Adding or removing the phosphate group can either activate or repress the protein
What does protein kinase do
adds phosphate group
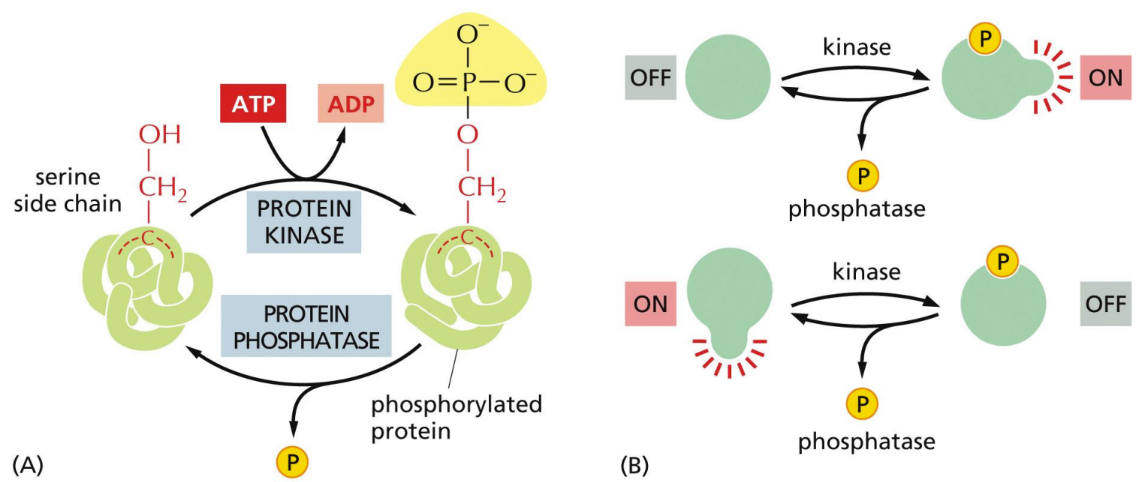
What does phosphatase do
removes phosphate group
What are motor proteins
to move molecules in or around cells
All work done by hydrolysis of ATP
EX:
Muscle contraction
Cell movement
Chromosomal movement during cell division
Moving neurotransmitter vesicles around neurons
Moving intracellular particles
Moves polymerases along a strand of nucleic acid
What is membrane transport
specific proteins pass through membranes (both cellular and organelle membranes) to act as channels or binders
Cell receptors = bound to cell membrane and catch extracellular signals
Have affinity for different things
Trigger a reaction inside the cell/organelle
Ex: glycoprotein cell receptors
What are some protein functions
Protein functions:
Tissue nutrition
Water distribution (balance)
Plasma buffer
Substance transport
Structural support
Antibodies, coagulation, hormones
Enzymes
Movement
Membrane receptors and transport
*PROTEINS ESSENTIAL FOR FUNCTION (too little or too much can result in disease)
What does abnormally folded proteins result in
Results in many disease because protein aggregates form due to a loss of cellular quality control
Normal proteins eventually begin to aggregate, forming amyloid fibrils
What is amyloid
when normal proteins begin to aggregate together because there are other proteins that are mutated (lost cellular quality control)
Cross-beta filament proteins (multiple sheets stacking together)
Particularly sensitive in the brain
Not ALL amyloid is bad
Are protease resistant
Ex: Alzheimer, type 2 diabetes, Parkinson, Huntington, rheumatoid arthritis, & cardiac arrhythmias
What is prion disease (PrP*)
An infectious agent that is comprised entirely of a misfolded protein (PRoteinaceous and Infectious viriON)
Hypothesized to infect and propagate by refolding abnormally into a structure that is able to convert normal molecules of the protein into the abnormally structured form (ALTER PROTEINS)
Induce amyloid
Able to convert normal PrP proteins into the infectious isoform (PrP*) by changing their conformation
Pulls other proteins/PrP to misshape with it
Ex: Mad cow disease, Creutzfeld-Jacob disease, & scrapie
What is Prion P (PrP)
a normal protein found on the membrane of cells (especially neurons)
Has 209 amino acids and is MAINLY an alpha-helical structure
the bottom box like protein is an amyloid
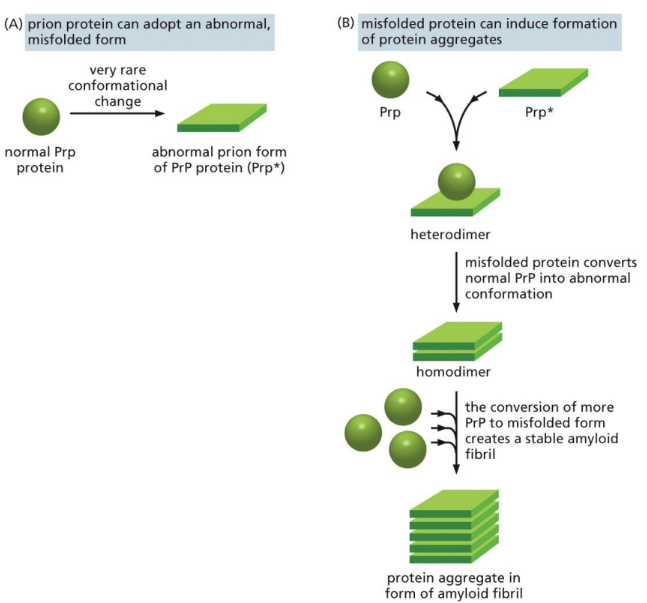
What are some prion related diseases
Mad cow disease, Creutzfeld-Jacob disease, & Scrapie
Results when normal PrP are converted into PrP*
What are CpG islands
surround housekeeping genes (need to be kept on all the time – important) that code for proteins that keep cells viable
Helps maintain cellular specialization
CpG methylation is important for efficient gene repression to be passed onto daughter cells
Surround the promoter
20,000 of CpGs mark the 5’ end of transcription units (typically around the promoter)
This ratio probably reflects a balance between methylated CG loss by DNA repair and CG gain by random mutation. The CG sequences that remain are very unevenly distributed in the genome; they are present at 10 times their average density in selected regions
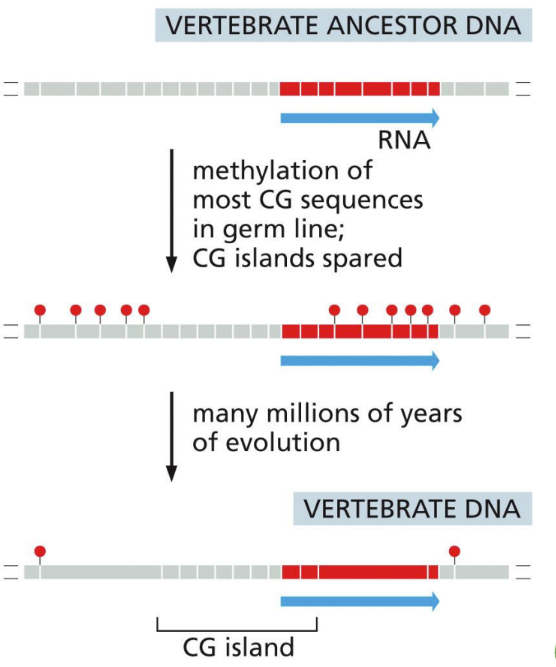
Epigenetic inheritance
heritable changes in phenotype (appearance) or gene expression caused by mechanisms other than changes in the underlying DNA sequence
Epi = in addition to => environmental
Can be affected by the environment
Ex: Lactose tolerance
LPH metabolizes lactose (encoded by the LCT gene on chr. 2)
LCT expression regulated by polymorphism in upstream gene MCM6 (if polymorphism is missing, then lactose intolerance happens)
LPH is high in European populations and lower in African populations => likely because they need more vitamin D than Africans
cis and trans epigenetic mechanisms
What are cis-epigenetic mechanisms
within the same DNA strand/molecule
DNA methylation
Histone modification
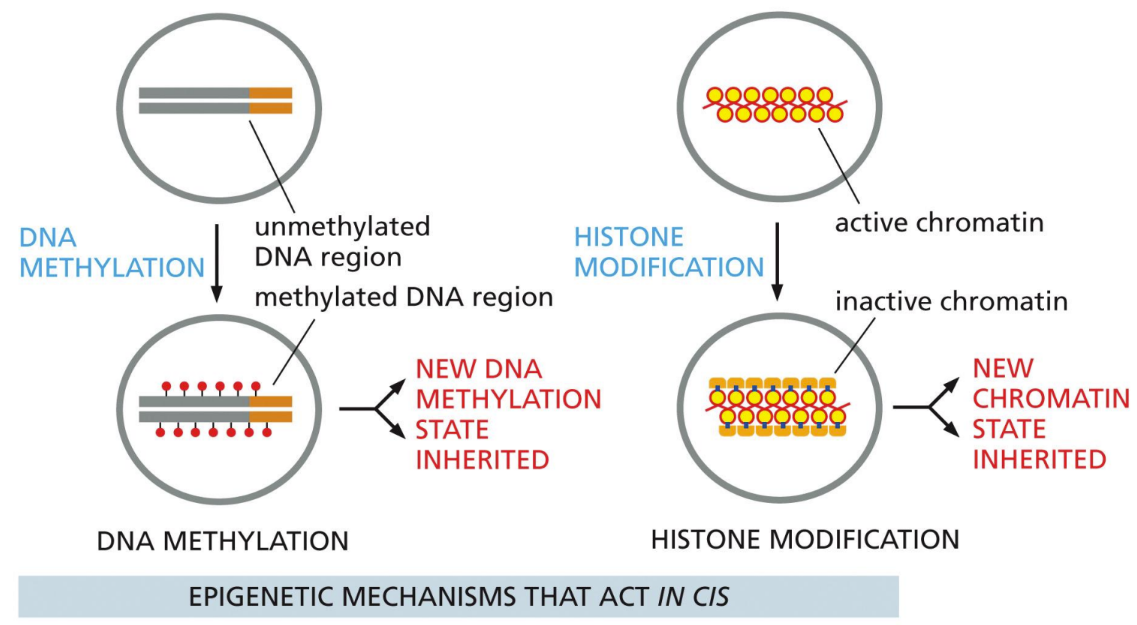
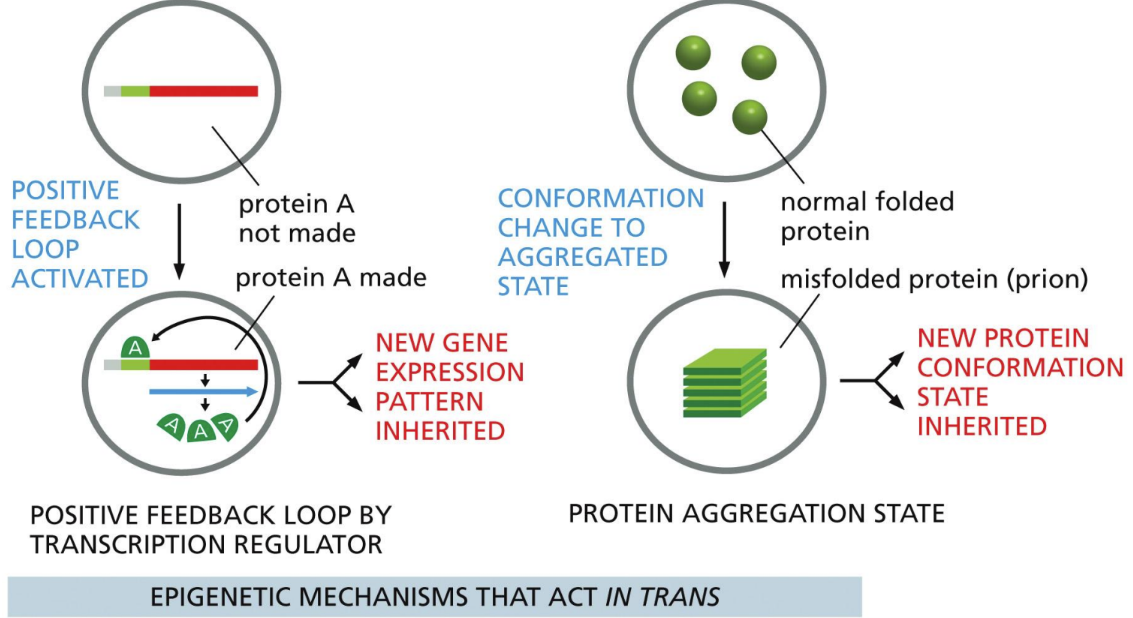
What are trans epigenetic mechanisms
something outside the sequence that’s affecting the gene
Positive feedback loop activated
Conformation change to aggregated state
What is imprinting
= silencing
A small minority of genes are expressed if inherited from mother, some expressed if inherited from father
Imprinting genes = one gene is silenced by methylation and cannot be transcribed
Normal inheritance = 2 working copies of a gene (one from each parent)
Imprinting DOESN’T cause a disease, the other remaining gene does
Disease: Prader-Willi & Angelman syndrome (imprinting of gene in chr. 15 & a deletion)
What is RNA interference (RNAi)
Short single-stranded non-coding RNAs of 20-30 nucleotides that guide or interact with other RNAs in the cell
Can inhibit translation of mRNA by catalyzing its destruction
Can mess with RNA and template during transcription and form repressive chromatin on the DNA
Proteins involved in RNAi are also involved in breaking apart dsRNA to form 23 nucleotide long siRNA
3 types (miRNA, siRNA, piwiRNA)
What are non-coding RNAs
lncRNAs = act as scaffold molecules (hold proteins together – after splicing)
Longer than 200 nucleotides
Ex: Telomerase RNA, Xist RNA, silencing RNA, gene regulators
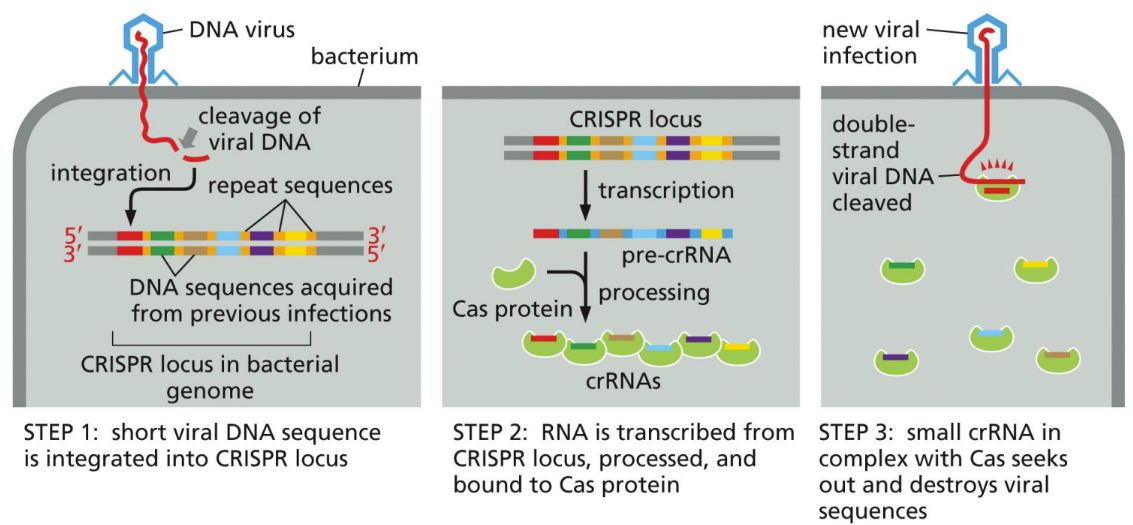
What is CRISPR
Viral DNA fragments are incorporated into bacterial genome, acting as a vaccine to make small noncoding RNAs (crRNA)
crRNAs destroy the virus if it reinfects the bacteria
Now using miRNA/siRNA (these are the Cas proteins that will interact with crRNA)
Cas proteins need a PAM sequence to recognize bacteria’s genome and cleave in the right place
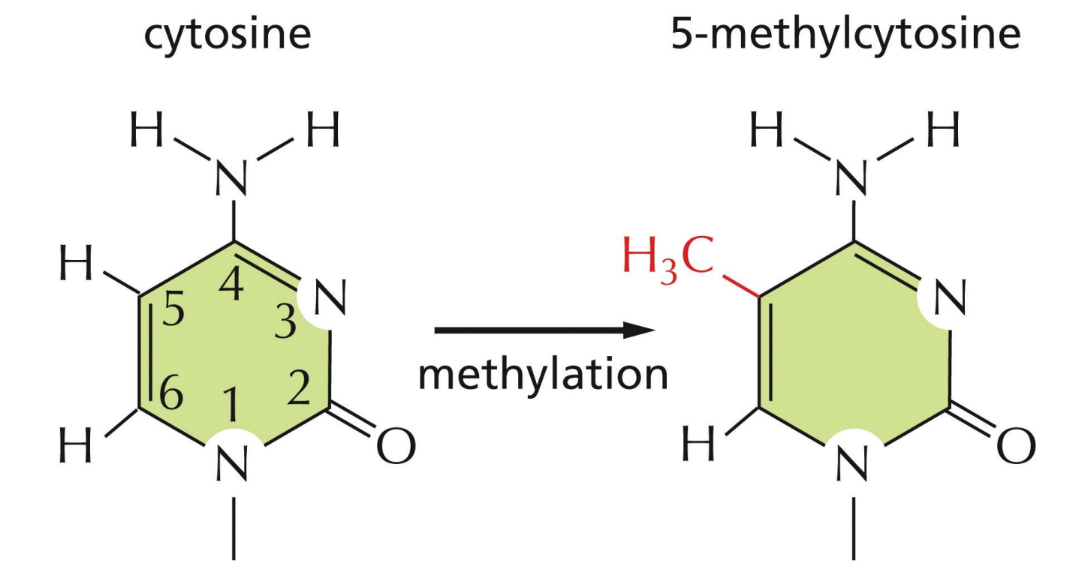
What does DNA methylation do
Methylation = methylating Cytosine
marks genes that are “imprinted”
Requires a methyl transferase to pass along the methylation to correct daughter cells
Regulate gene expression (without changing DNA sequence)
Methylation of CpG sequences is important in efficient gene repression that can be passed on to daughter cells
Helps maintain cellular specialization
Occurs right after DNA replication (inherited)
Methyl transferase recognizes methylated C at CpG and adds it to new daughter strand
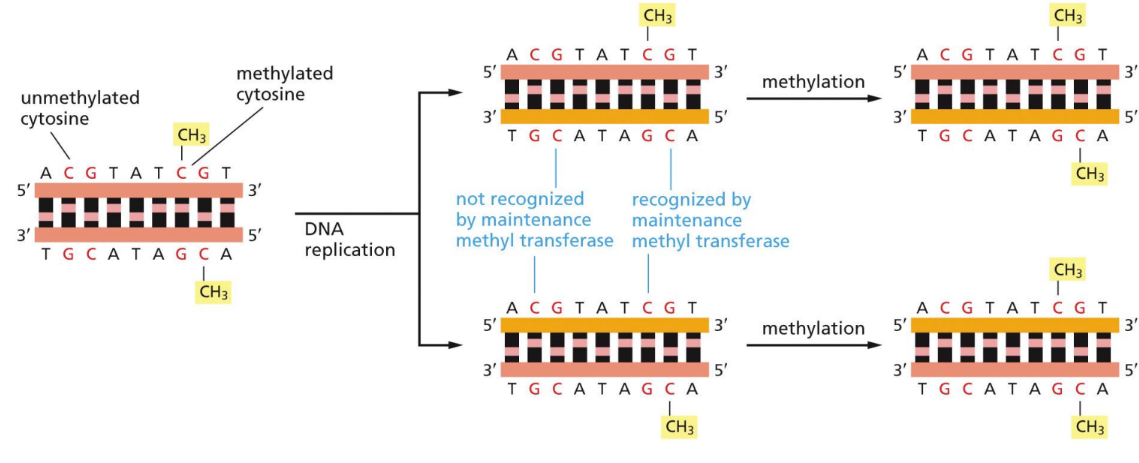
What is deamination
Deficient in mammals because the methylated cytosine mutate into a T over time (via accidental deamination)
The remainder, C, are present in CpG islands in selected regions of the genome
What are the 3 classes of RNAi
miRNA, siRNA, piRNA
What is micro RNA (miRNA)
Regulate some of human protein-coding genes by forming base pairs (complementary bp usually 7bp long in 3’ UTR of mRNA) with certain mRNAs & fine tune translation
Shut down method
Small but mighty
Regulate gene expression by repressing translation via mRNA degradation
miRNA calls other gene regulators to further reduce translation (mRNA of interest MUST have short common sequence in their 3’ UTR)
Made by RNA pol. II with a cap and poly-A tail. Proteins (Argonaute) are then added to form RISC (RNA-induced silencing complex)
RISC = proteins + miRNA complex
Involved in heterochromatin formation using methylated histones
What are small interfering RNA (siRNA)
Look for viral or transposable RNA element (transcriptional repressor) (defense mechanism?)
Self-replicating: RNA pols use siRNA as primer to make more precursor siRNAs
Broken apart and destroyed by RITS
Considered to be an ancient form of RNA interference
What are Piwi-interacting RNA (piRNA)
bind to piwi proteins and protect the germ line from transposable elements
Explain viral protection in relation to CRISPR
bacteria use non-coding RNA to destroy the DNA of invading viruses (via CRISPR)
Viral DNA fragments are incorporated into bacterial genome, acting as a vaccine to make small noncoding RNAs (crRNA)
crRNAs destroy the virus if it reinfects the bacteria
Now using miRNA/siRNA (these are the Cas proteins that will interact with crRNA)
Cas proteins need a PAM sequence to recognize bacteria’s genome and cleave in the right place
CRISPR is now used in plants and animals to manipulate genomes
What is Prader-Willi syndrome
aused by deletions of genetic region that includes small nuclear ribonucleoprotein polypeptide N gene
Maternal copy of gene is silenced (imprinted)
Paternal gene is deleted
Increased appetite (over-weight)
What is Angelman syndrome
Caused by a loss of expression of a single maternally expressed gene in the same region: UBE3A
UBE3A encodes for E3 ubiquitin ligase protein (involved in targeting proteins for degradation & is only imprinted in the brain)
Loss of UBE3A = abnormalities in normal protein degradation during brain development
The paternal gene imprinted
Maternal is loss
“smiling” all the time
What are transcription regulatory proteins?
recognize specific DNA sequences because of the surface features of the double helix (recognizes surface features)
Complex arrangements that respong to a variety of signals that are interpreted and integrated
Double helix features = major & minor groove
Binds to the major grooves because it has the cis-sequences & other contact points
Contain structural motifs
Don't need to unwind DNA to read sequences
In bacteria: transcription is controlled by 1 regulatory protein (and have operons)
In eukaryotes: many proteins control transcription & can act over very long distances
Single transcription regulatory proteins
Single proteins can make the final decision as to transcribe or not
Can form an entire organ by beginning a cascade of other proteins & cell-cell interactions
Ex: eyeless drosophila (eyes instead of legs form)
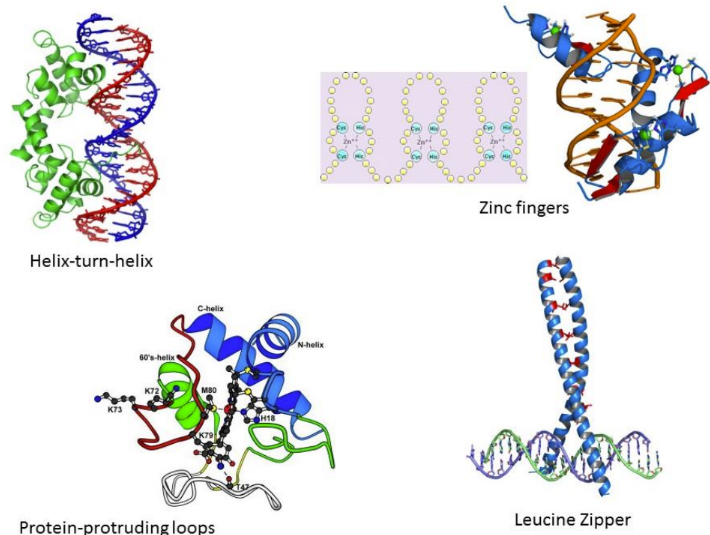
What are structural motifs
bind to the major groove based on the protein’s amino acid side chains (a recognizable, recurring 3D arrangement of secondary structure elements, such as helices and sheets, connected by loops)
Found in transcription regulators
Common motifs:
Helix-turn-helix = 2 alpha helices with a recognition sequence
Homeodomains are a type of helix-turn-helix
Zinc fingers = DNA binding proteins with a zinc (unique) moiety
zinc can hold alpha helix and beta sheet together
Zinc fills a two-alpha helix protein
Protein protruding loops =
Leucine zippers =
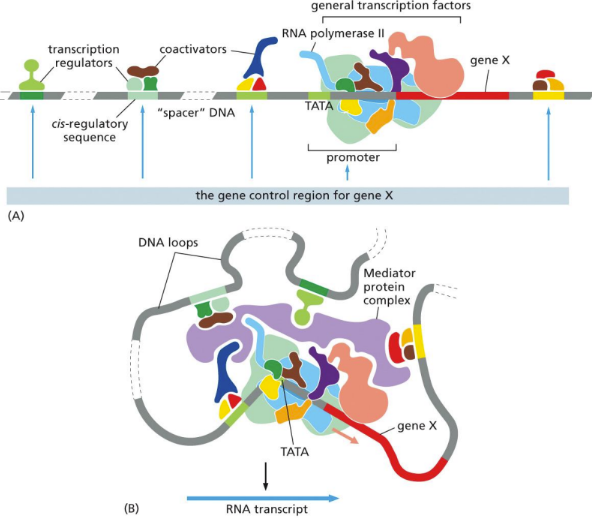
What are cis-regulatory sequence
DNA sequences specify the time and place that each gene is to be transcribed when read by their specific bound transcription regulators
DNA sequences recognized within the same molecule
Control transcription and are upstream of the transcription initiation start point
Positive regulators = activators
Negative regulators = repressors
Many protein regulators control a single gene (many regulator sequences within that gene
DNA looping critical for proteins, sequences, and polymerase to interact
“Complex switches”
What is the significance of DNA looping
DNA looping critical for complex switches to interact (it ISN’T LINEAR)
Brings activators/repressors and sequences closer to the mediator and other TFs
Gene control region (GCR)
consists of activator (and co-activator)/regulatory sequence/associated proteins, promoter region/factors/RNA pol, gene, more reg. proteins (chromatin remodelers)
Formed when a mediator is present and binds all the components together
RNA pol. II then transcribes all coding genes & non-coding RNA genes
How do cells control their own proteins? (list and describe)
Transcriptional control = DNA to RNA transcript
Controlled by DNA sequences (cis-regulatory sequences) near a gene upstream of the transcription initiation start point which are recognized by transcription regulator proteins
RNA processing
RNA transcript to mRNA
RNA transport and localization
mRNA being exported from the nucleus to the cytosol
Translation control
mRNA to protein
Degradation control (both mRNA & protein)
mRNA to inactive mRNA (degradation) or protein degraded into inactive protein
Protein activity control (phosphorylation)
Making a protein into active or inactive