MCB104 - Lecture 14 - Genome Annotation and Assembly & Genotypic Variation
1/82
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
83 Terms
What else needed on top of having a full genome sequence?
Full genome sequence - ONLY STEP ONE
We need to annotate the sequence to indicate what sequences promote what tasks
Without annotating, genome sequence is just a series of letters
What are Omics?
Collective characterization of complete sets of specific type of biological information, typically in high throughput manner
Genome: All DNA Sequences
Transcriptome: All transcribed RNAs
Exome: All Exons
Proteome: All Proteins
Metabolome: All metabolites
Connectome: All neural pathways
All these methods involve massive amounts of data, requiring various tools to filter out meaningful info
What is Euchromatin?
NOT Heterochromatin
What are Genomes?
Genome: All DNA Sequences
What are Transcriptome?
All transcribed RNAs
What are Exome?
All Exons
What are Proteome?
All Proteins
What are Metabolome?
All metabolites
What are Connectome?
All neural pathways
How can you identify genes through ORFs?
An open reading frame is a reading-frame uninterrupted by stop codons
DNA can be read in 6 reading frames (3 from each strand due to codon length)
Coding sequences tend to have longer ORFs

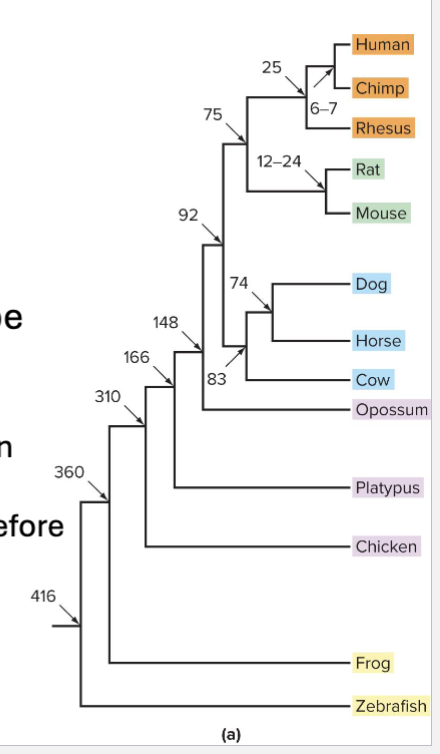
How can you identify genes through Phylogenetics?
Comparing genomes can help track natural selection
Phylogenetic trees depicting relatedness can be made by comparing DNA sequences among organisms
Branch points represent a series of nested common ancestors
Number at each branch point is millions of years before the present
What is Genome Conservation?
When comparing complete genome sequences, there are far less conservation than when only look at coding DNA sequences (CDS)
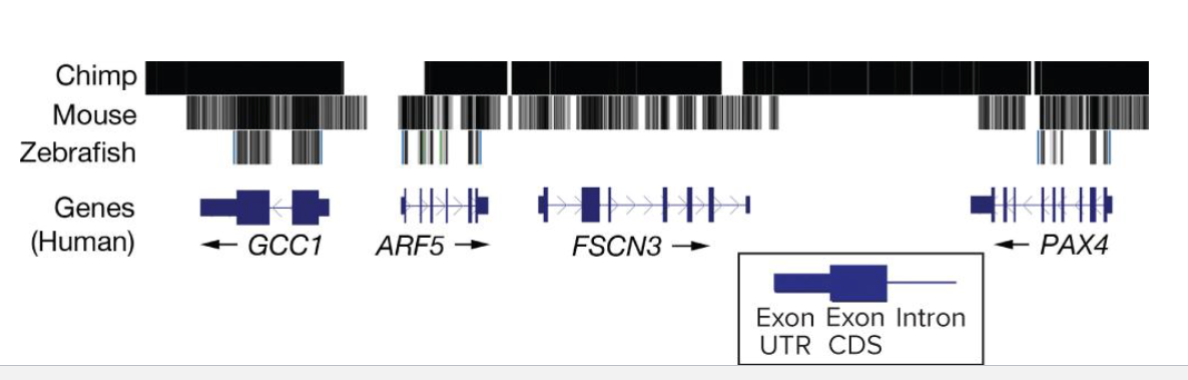
What is Visualizing Homology?
A homology map for a 100kb region of the human genome compared to 3 other species shows a range of overlap
Zebrafish overlap is largely contained to exons only, increasing overlap proportional to relatedness of species
What are Transcriptomics?
Sequencing RNA transcripts provides info about what sequences are actually transcribed
Directly sequencing RNA is difficult
Fragile/Unstable
Any given RNA sequence is pretty uncommon
Tech required is elaborate/expensive
Solution? CONVERT TO DNA
Use retroviral reverse transcriptase to copy RNA into more stable complementary DNA (cDNA)
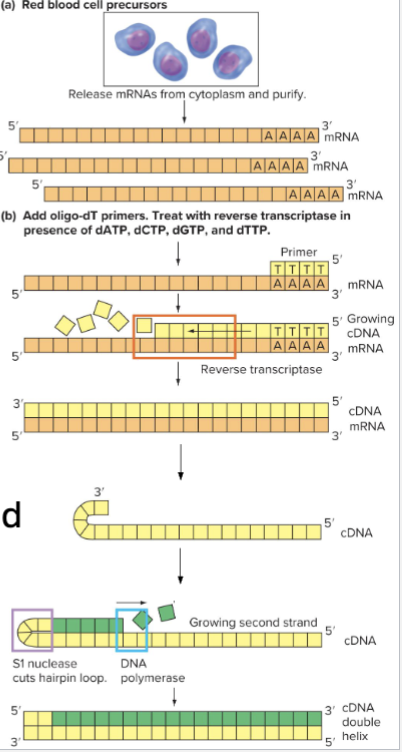
How can cDNA Library be created?
Isolate RNA from sample
Add primers
Use oligo-dT primers to bind to poly-A tails
Add Reverse Transcriptase and dNTPs
DNA Strand is synthesized
Denature and use RNase to degrade mRNA
Add DNA polymerase to synthesize second strand
Can self prime: 3’ end forms a hairpin loop
If used, need S1 nuclease to cleave loop after
Now have dsDNA sequences of mRNAs
How can cDNA library be preserved?
To preserve - clone it into vectors
Alternatively can be used as is for:
Next Gen Sequencing
Targeted PCR amplification
Many others
cDNA library includes only some exons
Only those transcribed in sample cells
Genomic library represents all regions of DNA equally, including introns
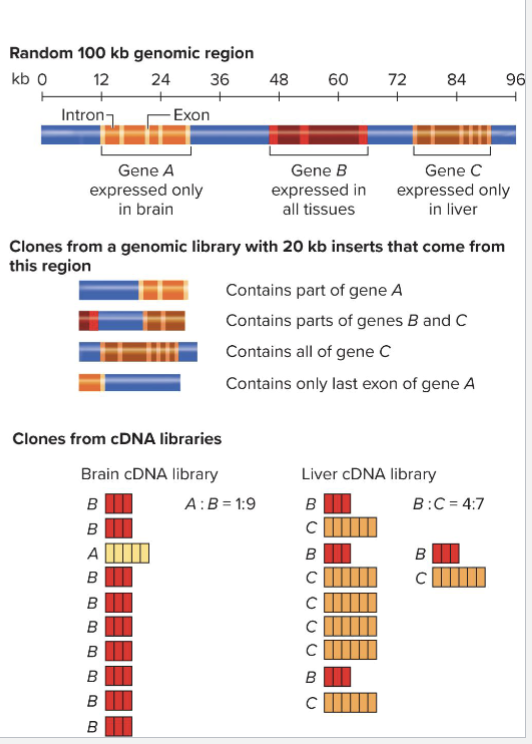
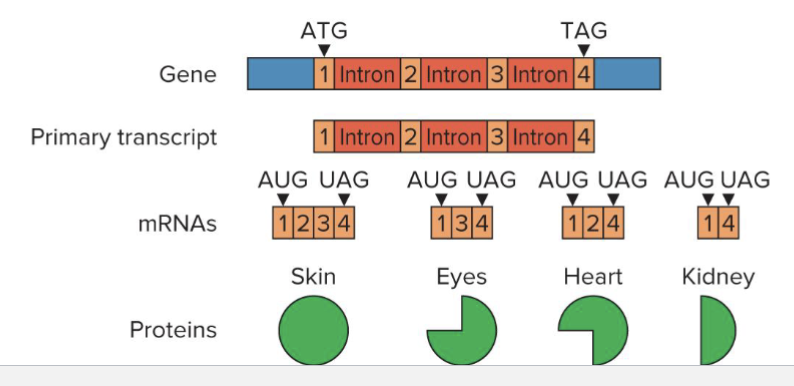
How does alternative splicing relate to cDNAs?
Alternative splicing complicates the prediction of proteome
Comparing cDNAs from different tissues can show where splicing occurs
Both “where in primary transcript” and “in which tissues”
What is the Genome Architecture?
The human genome has about 28,000 genes
Most DNA sequences are introns and other noncoding DNA:
Exome = 1.5% to 2%
Remainder is: introns, centromeres, telomeres, transposable elements, etc
Variation in genome size mostly due to changes in noncoding DNA rather than gene number or size
What are Repetitive Sequences?
Most of DNA outside genes is repetitive
Particular DNA sequences found many times in genome
2 types: multicopy tandem repeats and transposable elements
Repetitive DNA with no known function referred to ask junk DNA
Are Centromere/Telomere junk DNA?
NO
Centromeres anchor kinetochores
Telomeres protect the ends of DNA molecule and protect against replicative degeneration
What are gene-rich regions?
Chromosomal regions that have many more genes than expected from average gene density over entire genome
Example in human genome - class III region of major histocompatibility complex
What are gene deserts?
Regions that have no identifiable genes
Largest is 5.1 Mb on chromosome 5 with no identified genes
Describe gene-rich/gene deserts establishment with arrangement of genes
Biological significance of gene-rich regions and gene deserts isn’t well established
Both connected someway to gene regulation
What is MHC Region?
Class III regions of the human major histocompatibility (MHC) complex
Contains 60 genes within a 700 kb region
Most gene-rich region of human genome
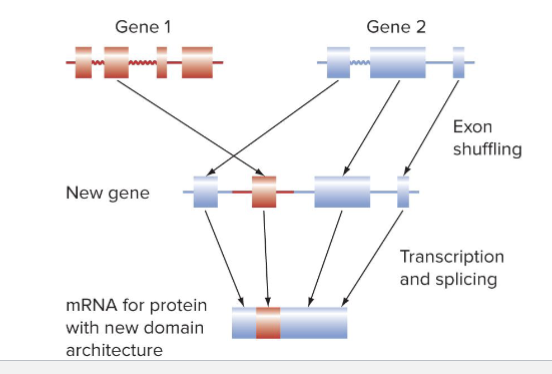
What is Genomic Evolution
Exons often encode protein domains: sequence of amino acids that fold into functional units
Shuffling, addition, or deletion of exons during evolution can create new domain architectures
Domain architecture: number, kind and order of protein domains
What is Domain Analysis?
Function of a new protein can be deduced if it contains a domain known to play a role in other proteins
Shown below: Homeobox Domain (found in Hox genes)

Can exon shuffling create new genes?
YES
After exon shuffling, protein products have novel domain architectures
Moving entire exons is more forgiving than moving parts of exons
Exons that have a specific function are more likely to stick when moved
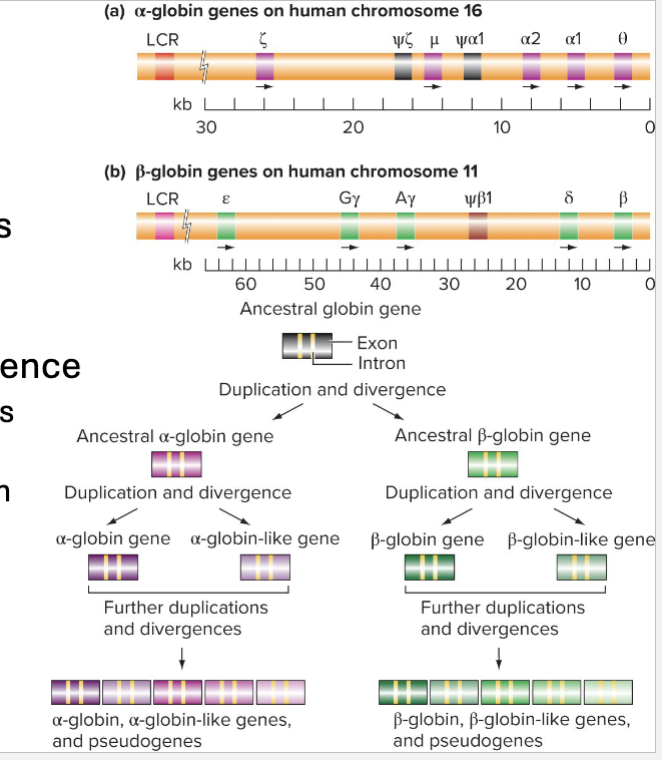
What are Gene Families?
Groups of genes closely related in sequence and function
Evolve via Duplication and Divergence
Duplicated DNA sequence products start out identical
Eventually diverge via accumulation of mutations
EX; Globin, Hox, Small Monomeric GTP (Rho, Ras, etc)
What is Homologous Genes?
Any evolutionary related sequences
What is Orthologous Genes?
Arose form same gene in common ancestor, usually retain same function
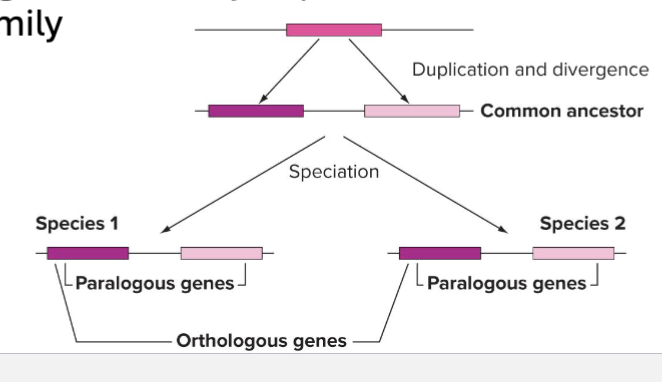
What is Paralogous Genes?
Arise by duplication, often refers to members of gene family
What are Psuedogenes?
Sequences that look like, but don’t function as genes
Rapidly accumulate mutations
Common features:
Missing promotor/start codon
Early frameshift/nonsense mutations
What are de novo genes?
Genes without homologs
Young genes that evolved recently from ancestral intergenic sequences
What are synthetic blocks?
Homologous blocks of chromosomal sequence
What are chromosomal rearrangements:
The cutting and reassembling of chromosomal blocks accompanying evolutionary divergence
How does Chromosomal Rearrangements relate to Mouse/Human genomes
Mouse and human genomes diverged 85 mya, but can be compared via chromosomes to visualize similarities
Is the number of genes in the genome the number of proteins the genome can generate?
NO, IT IS NOT
What does combinatorics and complexity relate?
Combinatorial amplification allow a comparatively small number of genes to produce a large variety of proteins
Remember product rule
Can occur at different levels
DNA level: The DNA itself is rearranged into different combinations
RNA level: Alternative Splicing
Protein level: Multimeric proteins and post translational modifications
What is VDJ Recombination?
Best studied DNA-level combinatorial amplification
T-cell receptors have Variable (V), Diversity (D), Joining (J) and Constant (C) segments
DNA rearrangement in T cell precursors combines V, D, and J segments into an exon
Done by deleting intervening sections
Result is about 1000 different combinations
Only occurs once per T-cell precursor (all other are gone, DNA itself is edited, happens only ONCE)
What is an example of Combinatorial Amplification: Alternative Splicing
EX: Neurexin Genes
2 alternative promoters; 5 sites for alternative splicing
3 different neurexin genes
Can generate >2000 different mRNAs
What is Bioinformatics?
The science of using computational methods to decipher biological meaning of information contained in organismal systems
What is GenBank (Bioinformatics)
Database established by NIH in 1982
Online repository of sequence data
What is RefSeq (Bioinformatics)
Single, complete, annotated version of species’ genome
Agreed upon standard for comparison
Maintained by NCBI
What is Basic Local Alignment Search Tool (BLAST)
Aligns query sequences with sequences in a database and finds areas of homology
Has variants for nucleotide and amino acid sequence searches
Amino acid search often includes information on how similar 2 amino acids are to improve alignment
Similar to conservative vs. nonconservative subtitutions
i.e: Leucine vs. Isoeucine Aspartic Acid vs. glutamic acid
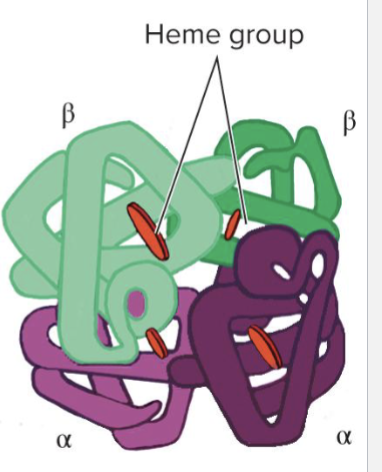
What is Hemoglobin?
Hemoglobin carries oxygen in blood
Adult hemoglobin consists of 4 polypeptide chains
2 alpha (a) globins
2 beta (b) globins
Each polypeptide chains surrounds a heme group that binds and releases oxygen
How is hemoglobin during development?
Embryonic (Z and e) and fetal (y) hemoglobins bind more tightly to oxygen to facilitate transfer of oxygen from mother to embryo or fetus
Adult hemoglobin binds oxygen less tightly to allow delivery of oxygen to organs
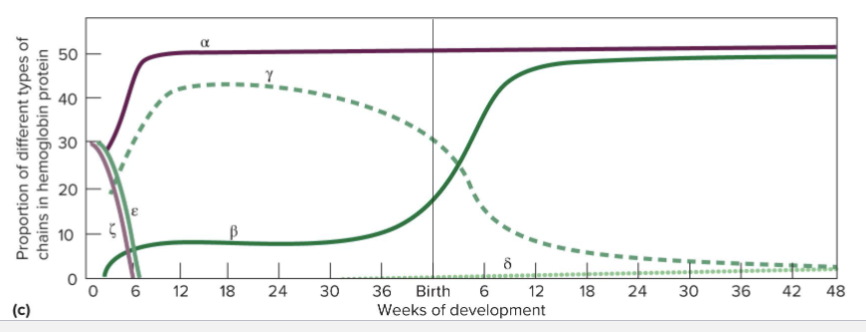
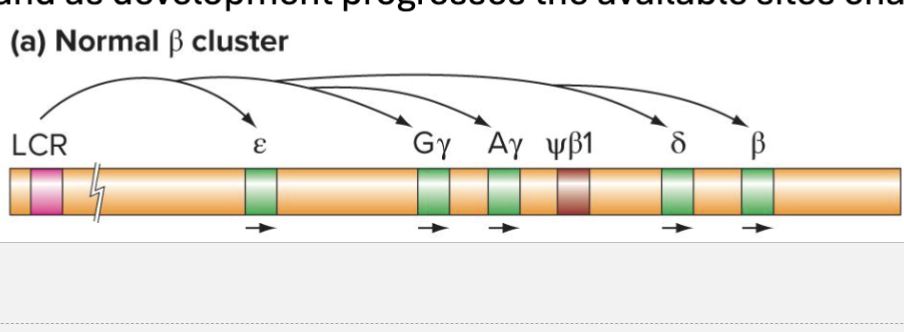
What is the Hemoglobin Cluster?
Globin Genes are in 2 clusters: (a and B)
Face same direction and are in order they are expressed during development
Clusters are controlled by locus control regions (LCR)
LCR are long range cis-regulatory elements with many enhancer sites
One model for function is that LCR forms loops to specific sites and as development progresses, the available sites change
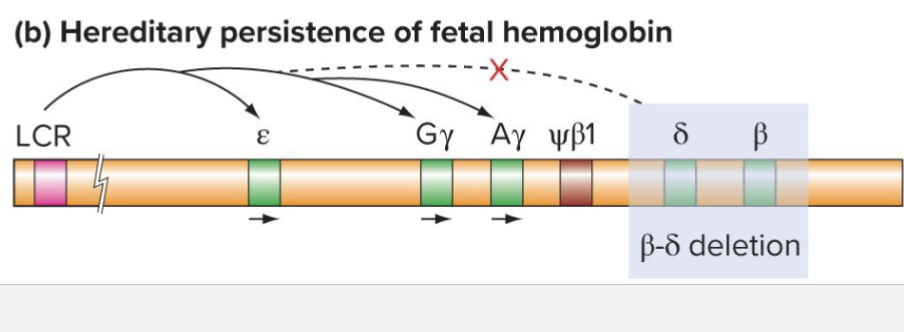
What is the hereditary persistance of fetal hemoglobin?
Rare condition of continued fetal globin expression
Deletion of omega and beta genes (should be lethal)
y genes continue to be expressed
Results in near normal level of health
LCR cannot switch to adult conformation as looping sites were deleted
What are globin related genetic disorders?
Hemolytic anemias
Changes amino acid sequence of alpha- or beta- globin chain
Causes destruction of red blood cells
Ex: Sickle Cell Anemia
Thalassemias
Mutations reduce or eliminate production of ½ globin polypeptides
Range of phenotypes
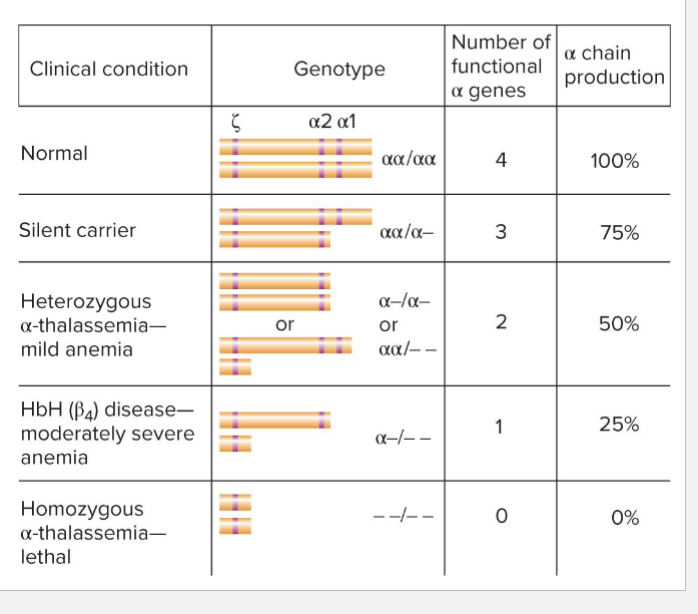
How are Thalassemias associated with alpha-globin deletions
Severity of thalassemia correlates with copy number
Requires at least 3 copies of alpha-globin to have normal blood
alpha 1 and 2 (a1 and a2) are relatively interchangeable
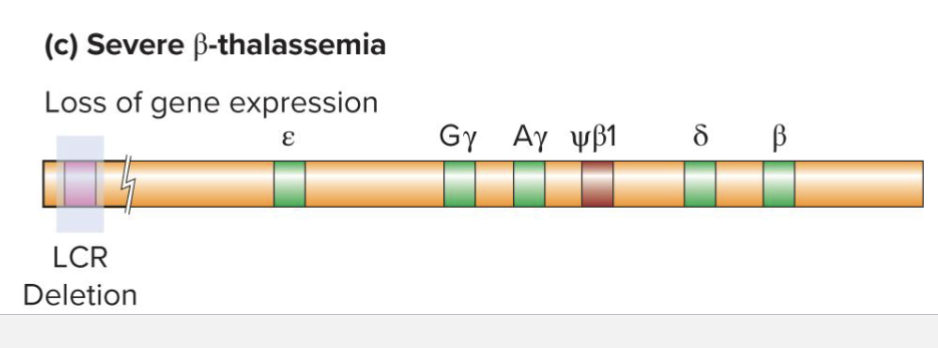
What is B-thalassemia?
Can occur via deletions of various B-globins however
Severe B-thalassemia occurs when LCR is deleted
Silences all B-globin genes even if all other regulatory sequences are preserved
Is there a “THE” human genome?
No,
The genome sequences of only 3 people reveal over 5.6 million DNA polymorphisms - sequence differences
Do most polymorphisms have a phenotype?
Codons make up less than 2% of human genome
Many mutations in codons don’t change amino acid
Many deleterious mutations disappear from population through natural selection
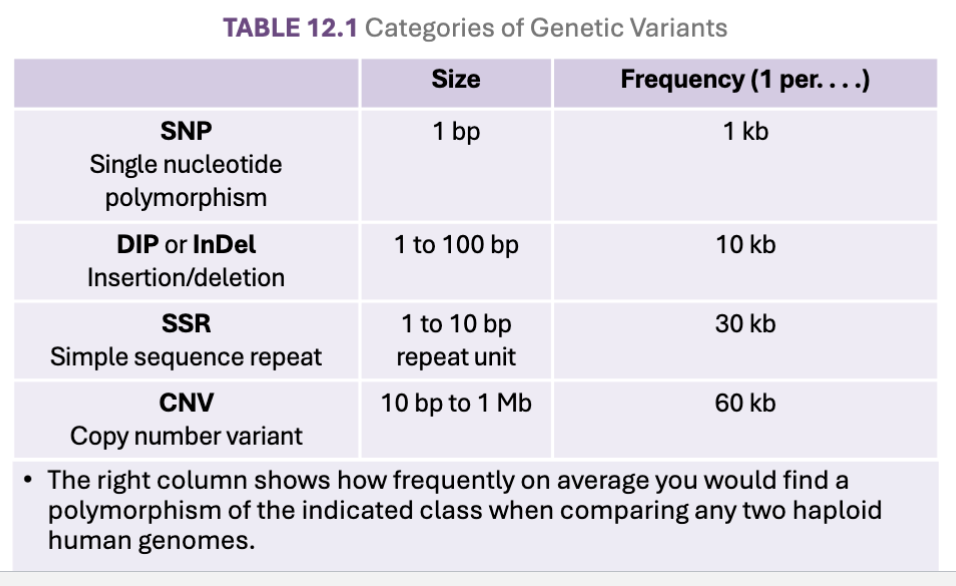
What are categories of variation?
Single nucleotide polymorphisms (SNPs)
• One base pair changes, the most common genetic variant
Deletion-insertion polymorphisms (DIPs or InDels)
Short insertions or deletions of a single or a few base pairs
In protein-encoding regions, DIP variants are frameshift mutations, unless a multiple of 3
Simple sequence repeats (SSRs or microsatellites)
1 to 10 base sequence repeated typically ~5-50 times in tandem, can rarely be >100
Most common repeating units are one-, two-, or three-base sequences
Copy number variants (CNVs)
Large blocks of genetic material up to 1 Mb in length that are variable in copy number in
the genomeMost important mechanism producing CNVs is unequal crossing-over in meiosis I
What are single nucleotides polymorphisms (SNPs)
Single nucleotide polymorphisms (SNPs)
• One base pair changes, the most common genetic variant
What are Deletion-insertion polymorphisms (DIPs or InDels)?
Short insertions or deletions of a single or a few base pairs
In protein-encoding regions, DIP variants are frameshift mutations, unless a multiple of 3
What are Simple sequence repeats (SSRs or microsatellites)?
1 to 10 base sequence repeated typically ~5-50 times in tandem, can rarely be >100
Most common repeating units are one-, two-, or three-base sequences
What are Copy number variants (CNVs)?
Large blocks of genetic material up to 1 Mb in length that are variable in copy number in
the genomeMost important mechanism producing CNVs is unequal crossing-over in meiosis I
What are the frequencies of Variations?
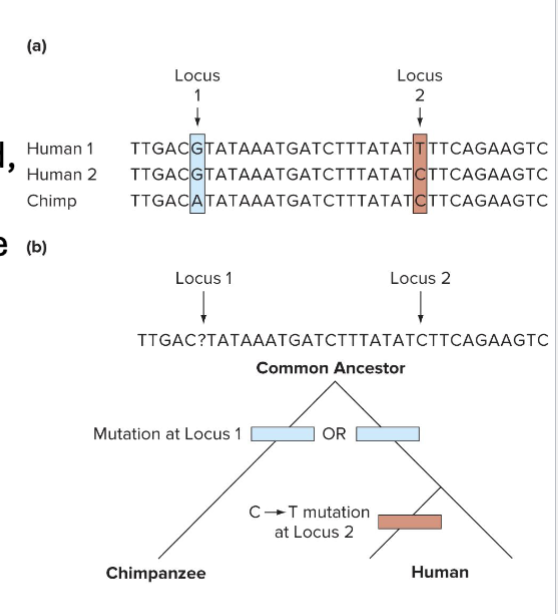
What is the Origin of Variations?
To determine when a variation occured, must compare genomes
Comparison of a human and chimpanzee genomes reveals the changes that has occured since divergence of these species
The single base change at loci 2 is polymorphic in humans
C is ancestral, present in ancestral organisms
T is derived, changed nucleotide
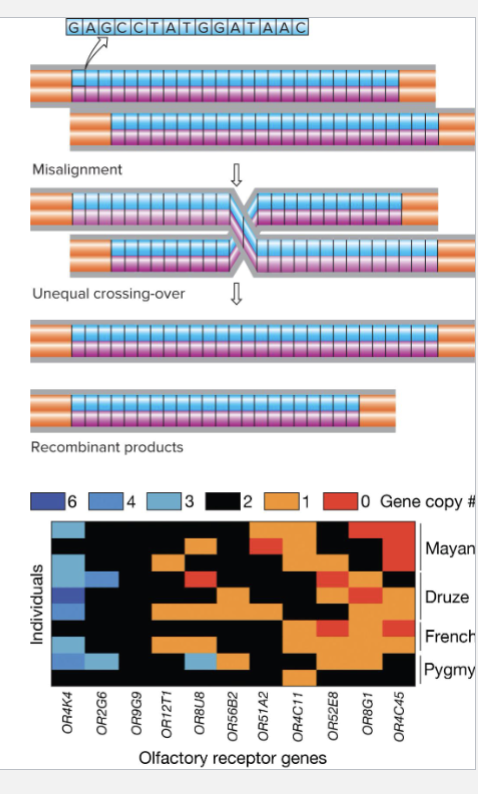
What are CNVs?
CNVs are tandem sequence repeats more than 10bp long
Misalignment during meiosis leads to unequal crossing over
Not common event, so most CNVs are inherited rather than being a new mutation
Example: Humans are fewer than 1000 olfactory receptor genes are different loci
At each locus, copy number varies
What is an overview of PCR?
Polymerase chain reaction (PCR)
Method of making many copies of target region of DNA
First developed in 1985
Fast and extremely efficient: can amplify DNA from single cell
Hinges on thermostable DNA polymerase
Originally Taq
Exponential increase in targeted DNA
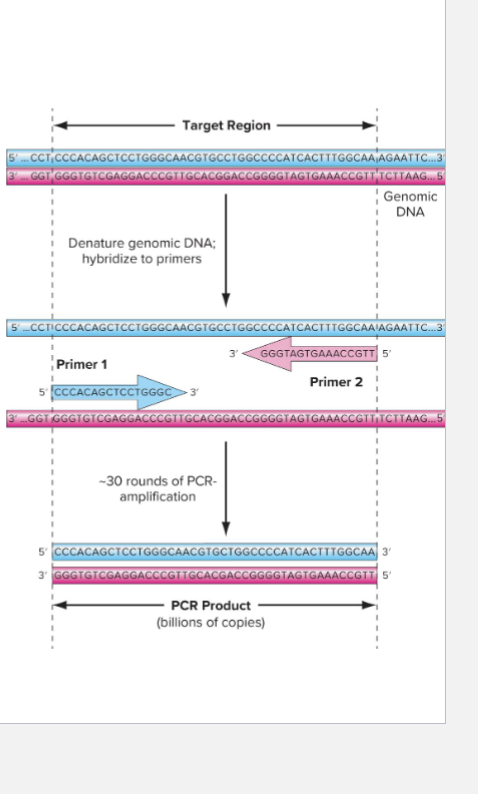
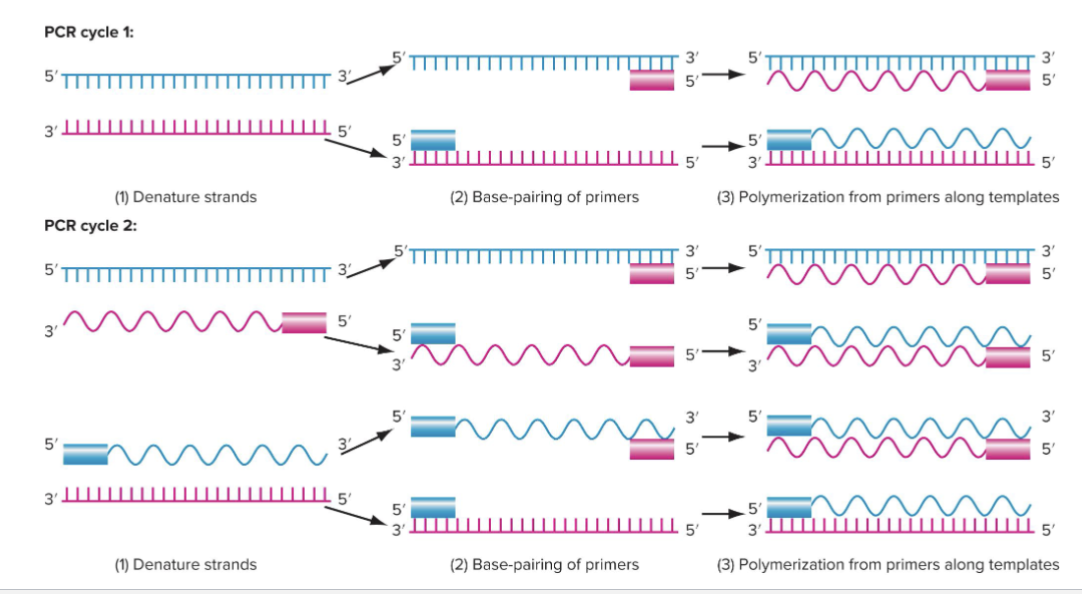
What are the steps of PCR?
CYCLE 1
1) Denature strands
2) Base-pairing of primers
3) Polymerization of primers along templates
CYCLE 2
1) Denature strands
2) Base-pairing of primers
3) Polymerization of primers along templates
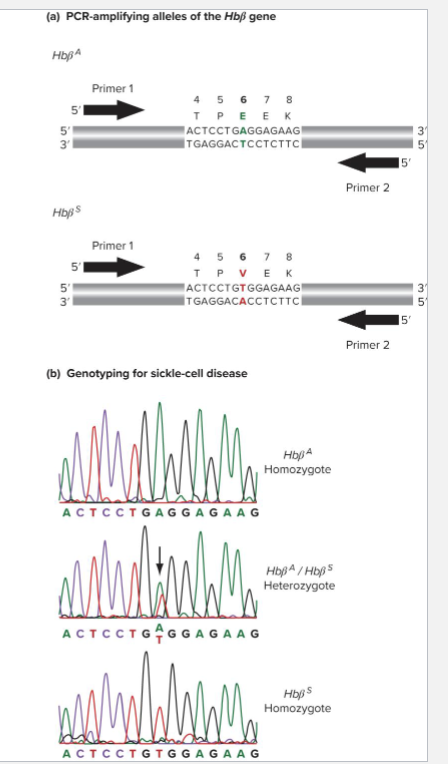
How does Genotyping via Sequencing work?
PCR amplify a targeted sequence
Use same primers or nested one to sequence allele
Usually Sanger Sequencing variant
EX: Sickle Cell anemia is caused by SNP in Hbbeta gene (Hbb)
Genotyping can idetify carriers and homozygous individuals
How does Genotyping by PCR product size work?
Amplify target sequence
Run PCR product on gel
If alleles have different length then PCR products with those alleles will run at different speeds
Can distinguish between homo and heterozygotes
VERY EASY TO DO YAYYYYY
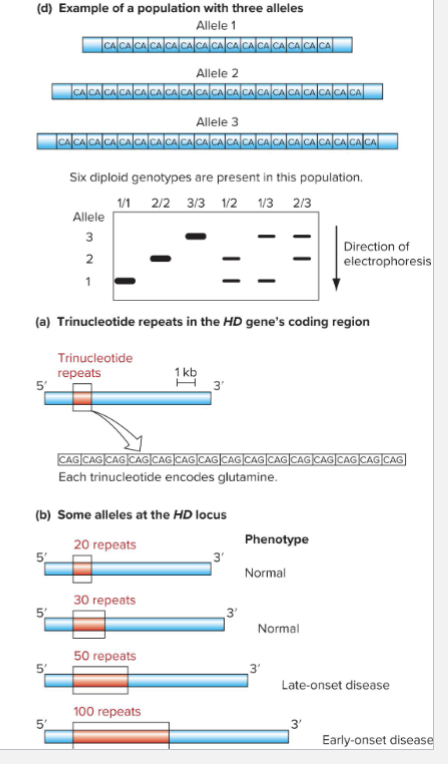
What is PCR Product Size Variations?
Size variations can be detected by gel electrophoresis
Ex: Huntington disease locus
Normal allele has <36 C A G repeats
Disease-causing alleles have 36 or more CAG repeats; alleles with 42 or more repeats are completely penetrant
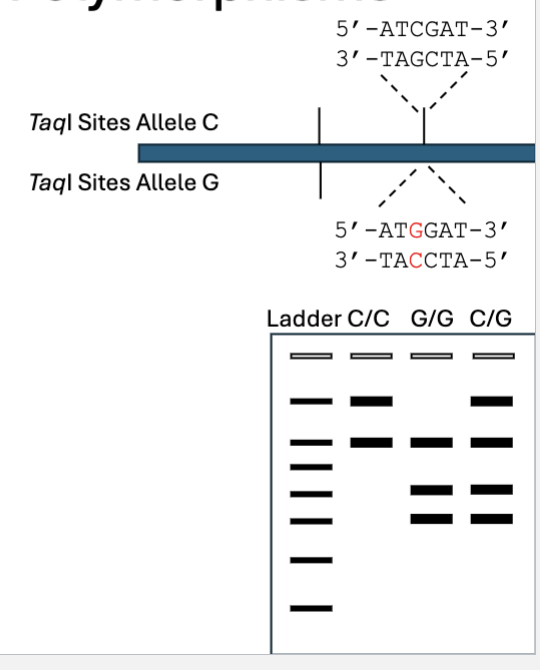
What are Restriction Fragment Length Polymorphisms (RFLPs)
Amplify target region with PCR
Digest with restriction enzyme
If variant disrupts/creates a restriction site, can distinguish between them
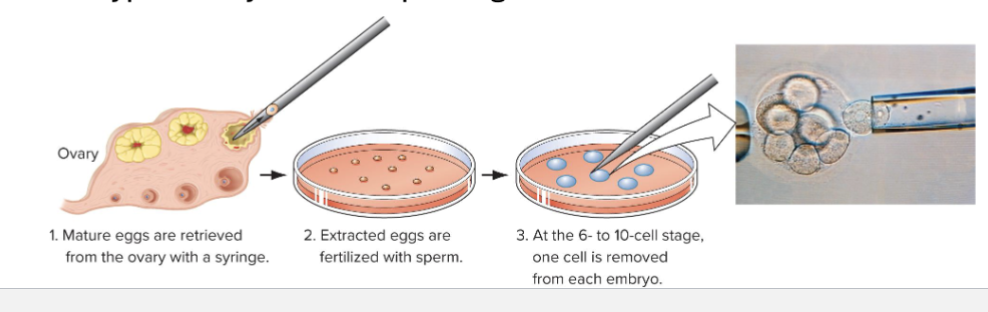
How can Fetal and Embryonic cells be genotyped using PCR
Prenatal genetic diagnosis
Genotyping fetal cells isolated by aminocentesis: fetal cells in amniotic fluid are extracted using a needle
Preimplanatation embryo diagnosis
Utilizes in vitro fertilization and PCR
Genotype embryos before placing in womb
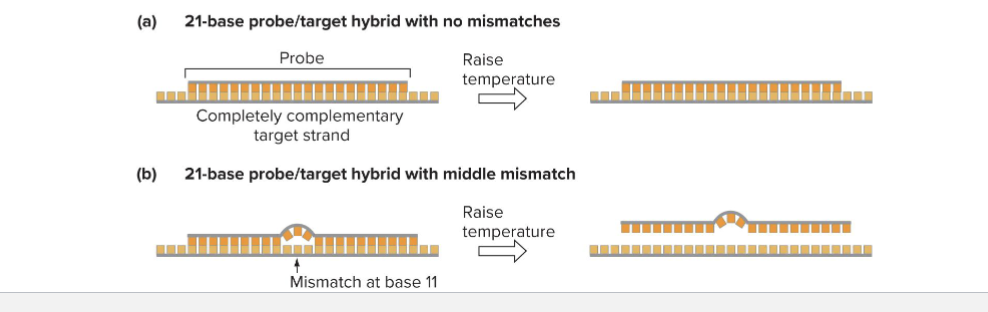
What are Hybridization Probes?
Hybridization of short (<40 bases0 oligonucleotides to sample (target) DNAs (allele-specific hybridization)
If no mismatch between probe and target, hybrid will be stable at high temperature
If mismatch between probe and target, hybrid will NOT be stable at high temperature
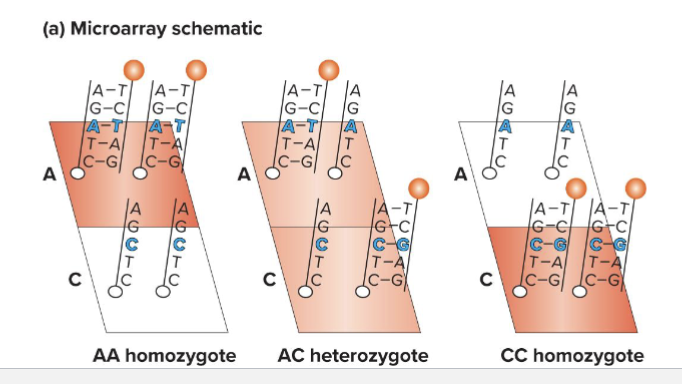
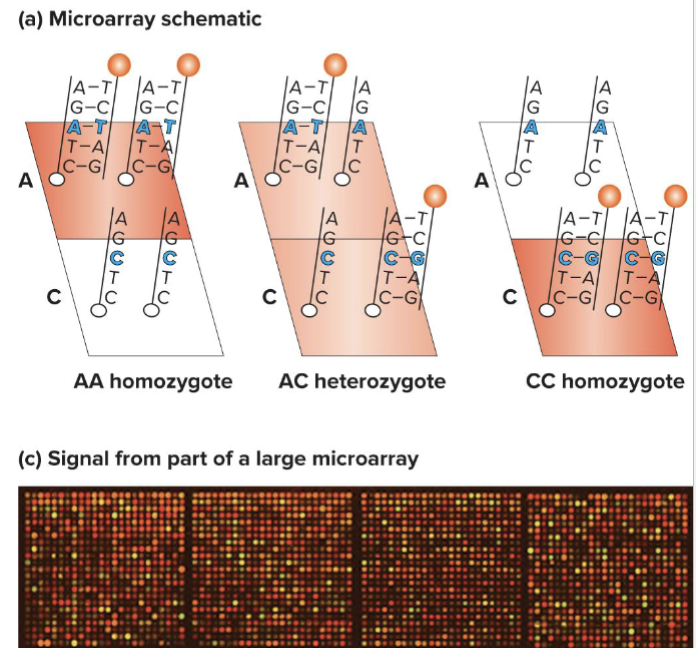
What are Microarrays?
Allele-specific oligonucleotides ( A S Os) are attached to a solid support (like silicon chip)
2 oligonucleotides are shown here, but many can be put on one array
How does microarray for genomic DNA work?
Preparation of genomic DNA for microarray
Fragmented
Adapter attached
Amplified by PCR denatured to make single stranded
Fluorescent dye coupled to end of single stranded DNA
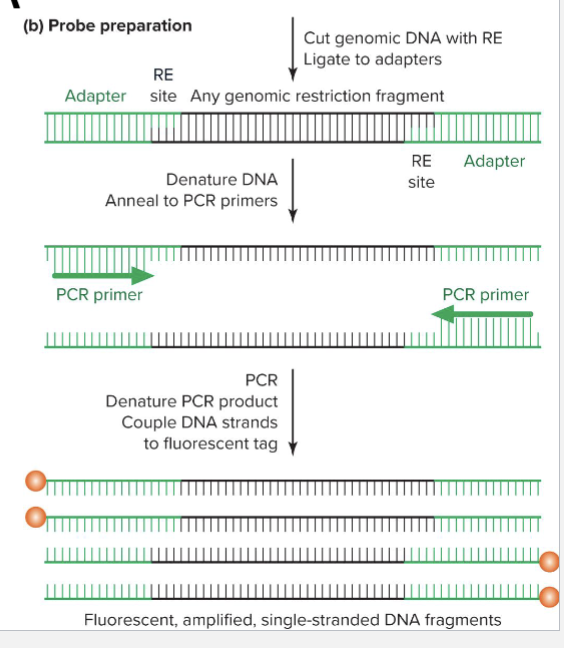
How can a microarray be read?
Fluorescent output is proportional to number of copies of each allele
Can sometimes distinguish between hets and homozygous
Up to 4 million loci can be genotyped simultaneously for approximately $100
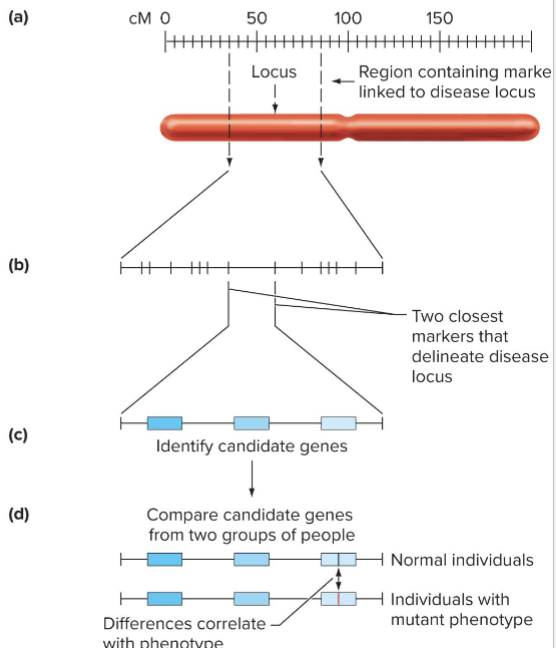
How does Positional Cloning work?
Positional Cloning
Object is to identify disease causing genes by genetic linkage to polymorphic loci
Strategy
Same as linkage analysis using 2 phenotypes, except one gene tracked by phenotype, the other by DNA genotype
Use microarrays to simultaneously analyze millions of 2 point crosses, each one a test for linkage between a disease locus and DNA marker
What are steps of positional cloning?
Region of interest narrowed by finding closely linked DNA markers
Candidate genes are located in region of interest
Sequence and expression of candidate genes are determined in normal and diseased individuals
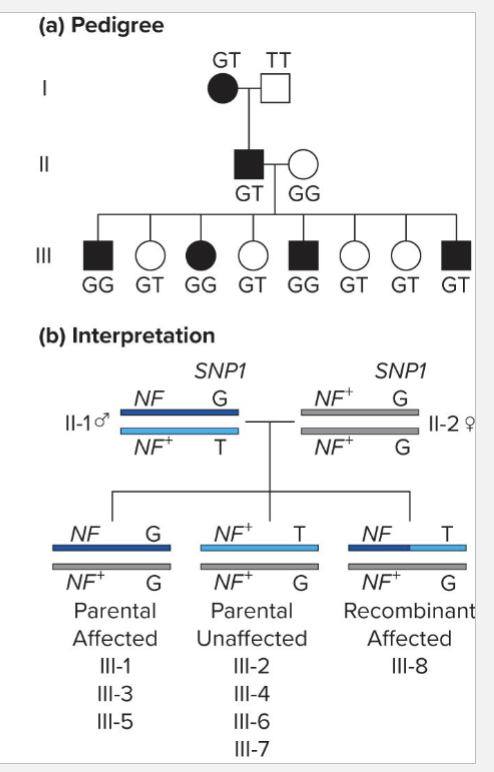
What is an example of using positional cloning?
NEUROFIBROMATOSIS
Autosomal, dominantly inherited
Causes proliferation of nerve tissue forming tumerous bumps
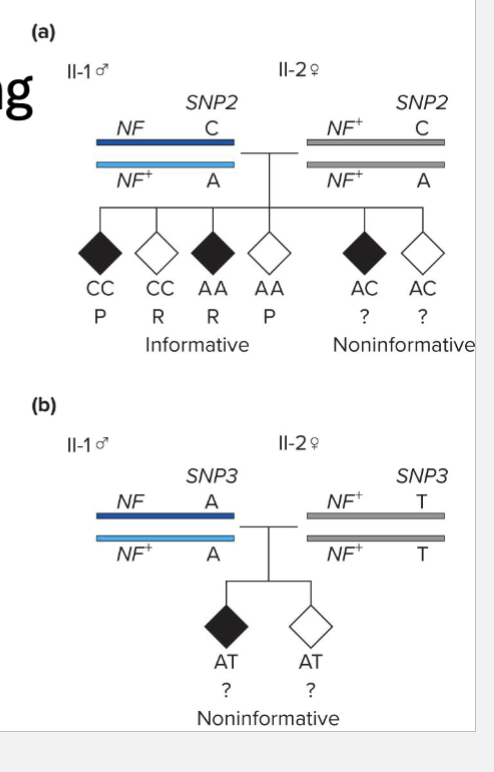
Positional cloning example determines whether or not SNP is linked to neurofibromatosis gene
Children in GEN III are in effect the result of testcross
RF = 0.125 (N too small to be confident though)
What are some limitations of positional cloning?
Configuration of alleles are not always known
Not all mating are informative
Difficult to obtain pedigree data in humans
Identified SNPs aren’t necessarily causative
How are large pedigrees used for positional cloning?
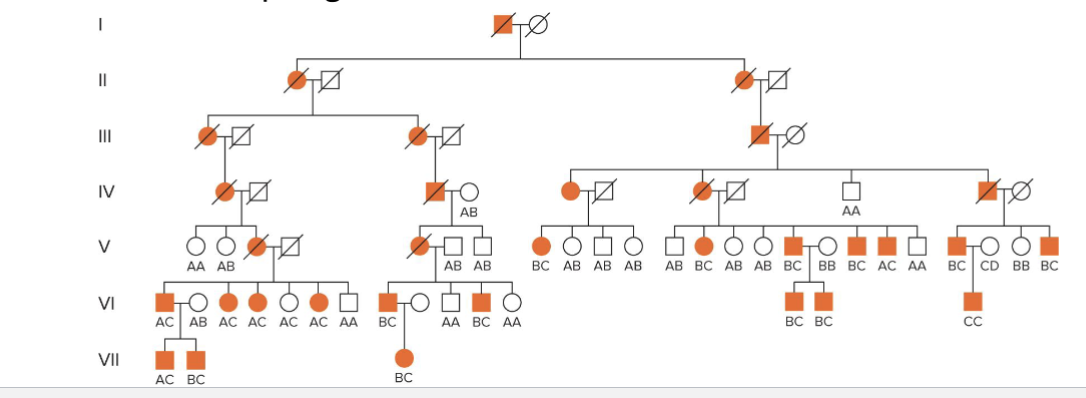
Mapping of Huntington disease
Detection of linkage between DNA marker G8 and HD locus
Segregation of G8 DNA marker (4 alleles - ABCD) in a large Venezuelan pedigree affected with HD
What is Lod Score?
Lod Score (Log of the odds) is used to determine if data is sufficient to conclude with confidence that a disease gene and a marker are linked
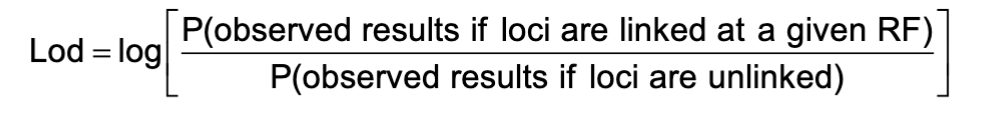
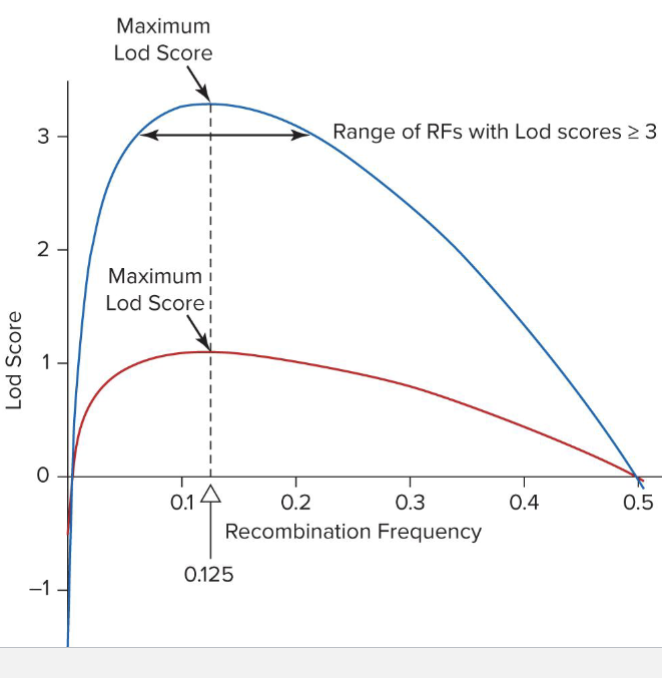
What is Lod Score compared to RF? Relationship?
Relationship between Log Score and assumed RF
Lod of 3 means log1000 so 1000x more likely to be linked than not
Can add Lod scores from different pedigrees together to increase confidence
RED: Lod score from Neurofibromatosis pedigree
BLUE: sum of Lod scores from 3 such pedigrees
Describe Locus heterogeneity:
Allelic heterogeneity: disease caused by different mutations in same gene
Compound heterozygotes (trans-heterozygotes) - individuals with different mutant alleles of the same gene
Individuals with certain alleles may respond to drug treatment, while others do not
Locus heterogeneity: disease caused by mutation in one or two more different genes
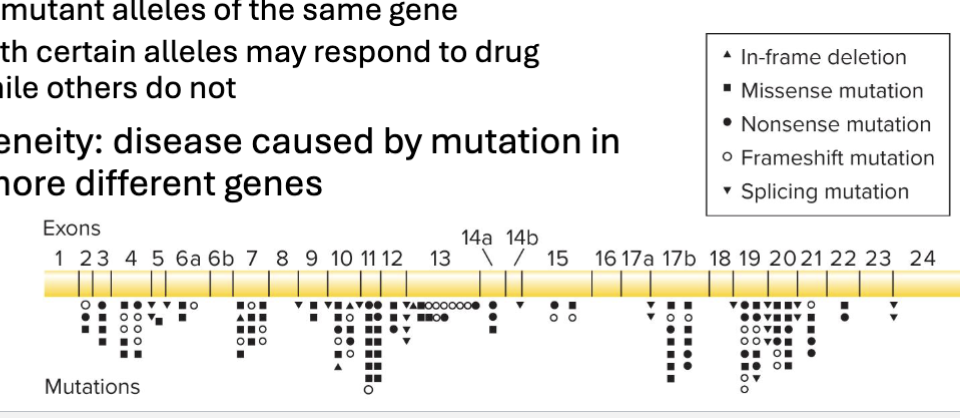
Describe an example with completely sequenced genomes
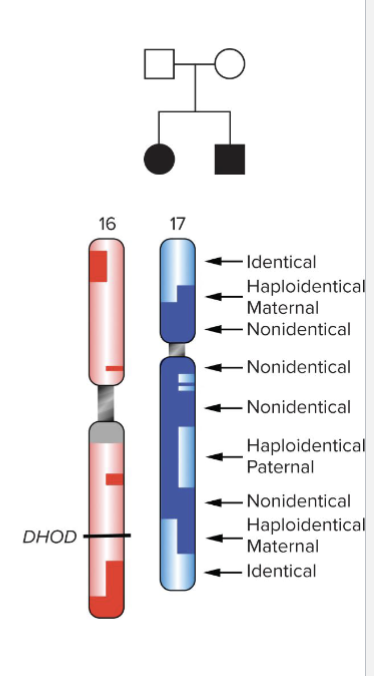
MILLER SYNDROME - First sample with completely sequenced genome
Pedigree:
Identical regions: brother and sister share same alleles
Nonidentical: siblings share NO alleles
Haploidentical maternal: same allele from mother, different from father
Haploidentical paternal: same allele from father, different from mother
Geneticists studying disease in affected children could focus on identical regions
Identified compound heterozygote in DHOD
How can Gene location be narrowed down?
Identifying causative alleles via filtering
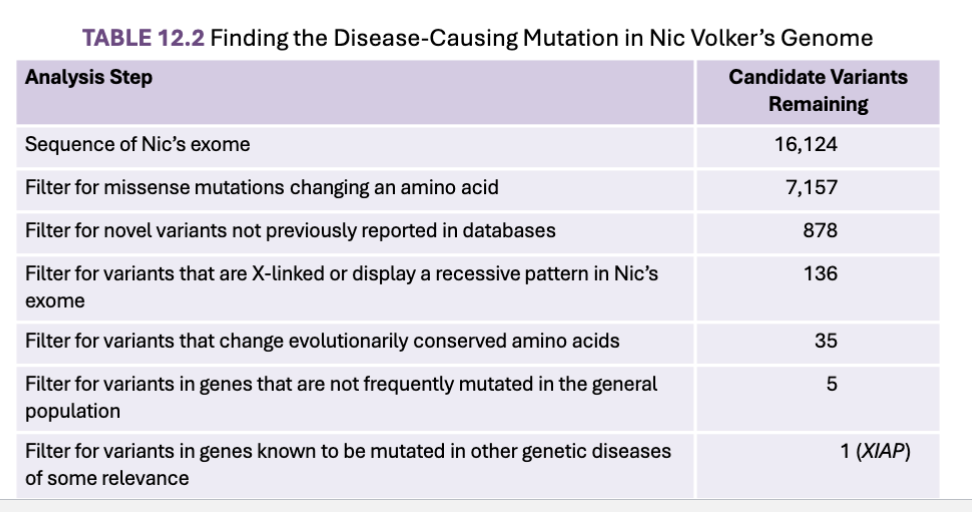
What are evolutionary conserved amino acids?
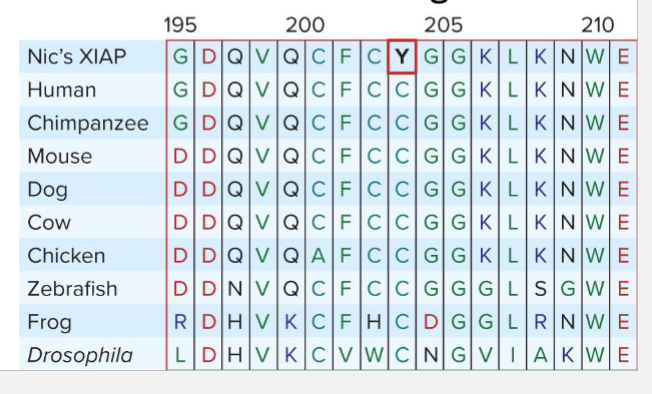
Nic’s XIAP gene had a missense mutation that changed a single amino acid that was completely conserved among humans, frogs, flies, and other species
Assumption that nonconversative variations in conserved regions are more likely to be causative