3L4-5 Prokaryotic Transcription
1/57
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
58 Terms
mRNA
Messenger RNA (mRNA) contains sequence of bases that encodes the primary amino acid sequence for a protein.
A mRNA serves as the template for translation by a ribosome.
tRNA
Transfer RNA (tRNA) carries an amino acid into the catalytic site of a ribosome.
The tRNA base pairs to mRNA to ensure the selection of the correct amino acid for incorporation into a nascent polypeptide chain.
rRNA
Ribosomal RNAs (rRNA) are structural components of a ribosome, the enzyme that catalyzes translation.
ncRNA
Noncoding RNAs (ncRNAs) do not encode proteins but have a variety of catalytic, structural, and regulatory functions.
gene
DNA encoding a protein + DNA regulatory elements
primary transcript
bacteria = DNA > primary transcript / mRNA = ready for translation
eukaryote = DNA > primary transcript > modified into final mRNA = ready for translation
ORF
open reading frame, start codon > end codon
continuous sequence that encodes primary sequence of a protein
“cistron” or “coding sequence”
cis v trans acting factors
trans-acting factors = diffusible, DNA-binding proteins (TFs)
cis-acting factors = fixed places tied to genes, only impact their gene, are close
transcription factors
DNA-binding proteins, trans-acting factors
transcription is catalyzed by
RNA polymerase
transcription starts at ___, ends at ___.
promoter (+1);
terminator
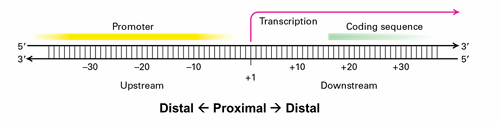
regulatory elements are located
upstream
proximal v distal
close;
far
RNA polymerase mechanism
similar to DNA polymerase
RNA polymerase can start synthesizing RNA de novo (without a primer )
Substrate is ribonucleoside 5’triphosphates (NTP)
Forms phosphodiester bond between 2 NTPs to begin RNA synthesis
The 5’ end will contain 3 phosphate groups
requires template DNA
no proofreading exonuclease activity
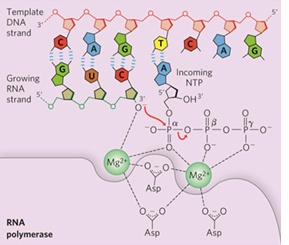
template v nontemplate
noncoding v coding

promoter sequence defines
template v nontemplate
prokaryotic RNA polymerase
e coli has only one
core enzyme and holoenzyme
RNA polymerase core enzyme
carries out transcription elongation
subunits a2 are essential for enzyme assembly and interact with activators
Subunits β and β’ form the catalytic core
Subunit ω provides structural stability
RNA polymerase holoenzyme
initiates transcription and synthesis of first 10 NTs
subunit sigma recognizes a promotes
stages of transcription
Initiation
Rate of transcription initiation is the major determinant of gene expression
Separate DNA strands
RNA polymerase holoenzyme
Elongation
RNA polymerase core enzyme
Synthesizes RNA 5’→3’
Moves along template strand 3’→5’
Termination
RNA polymerase core enzyme reaches terminator sequence to stop transcription
prokaryotic promoters
located on -35 and -10 regions;
RNA poly sigma subunit recognizes and binds, bending DNA to open it
consensus sequence
certain NTs are very common at each position
sigma’s consensus sequences
-35 = 5 TTGACA 3
-10 = 5 TATAAT 3
UP
upstream promoter element;
third AT rich recognition element, -40 > -60, highly expressed genes
bound by alpha subunit of RNA polymerase
constitutive promoters
active always, used in housekeeping genes
rate of transcription initiation is determined by __
similarity to bacterial promoter consensus sequence
strong v weak promoters
high sequence similarity with consensus;
several base differences
transcription initiation
holoenzyme binds to promoter = closed complex
unwinding DNA 12-15 bp > transcription bubble = open complex
holoenzyme initiates RNA synthesis and synthesizes 10 NTs
closed complex
holoenzyme + promoter
sigma 70 identifies promoter and makes protein-DNA interactions
transcription elongation
sigma70 dissociates > core enzyme
RNA polymerase completes promoter clearance
NusA protein replaces sigma70
transcription elongation rate increases
translation begins before transcription finishes
NusG binds RNA poly and ribosomes link complexes
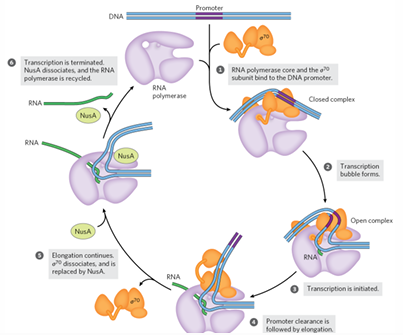
transcription elongation rate can be slowed by
RNA secondary structures
NusG
binds RNA poly and ribosomes link two complexes
rho (p) independent termination
terminators have hairpin region in RNA
long repeat of U, DNA has lots of A (U-A = 2 H bonds, weak)
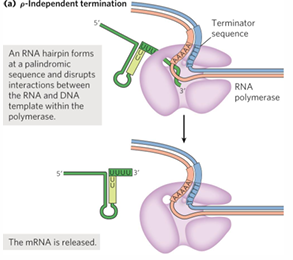
rho dependent termination
rho-dependent terminators have CA-rich sequence called rut (rho utilization) element
rho binds the rut in RNA, promotes release of RNA
rho
protein factor with ATP dependent RNA-DNA helicase
rut site
Rho UTilization site, rho binds to and promotes release of RNA
operon
coordinately regulated gene clusters, arranged as transcription unit;
some have constitutive promoters
prokaryotic
polycistronic mRNA
formed with one promoter and terminator,
has multiple ORFs encoding a different protein
inducible promoters
gene regulation changes to environment, it can be turned on and off
specificity factors
alter specificity of RNA poly for given promoter / set of promoters
repressors v activators
enhance RNA polymerase-promoter interaction
transcription factors are ___acting elements
trans
predominant structure in e.coli is
helix turn helix motif
recognition alpha helix
makes sequence-specific interactions
motif causes alpha helix to stick out from protein,
interacts with bps through grooves, forms H bonds and VDW with bps
exposed base pair chemical groups
each base pair presents a unique set of chemical groups in the major groove
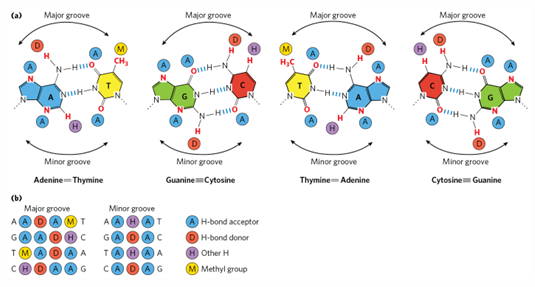
major groove v minor groove specificity
major can always specify AT v TA v GC v CG
minor groove can only show AT/TA v GC/CG
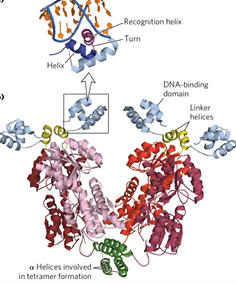
helix-turn-helix motif
predominant DNA binding domain, defined by 2 alpha helixes.
second helix is the recognition helix
bacterial TFs
most bind DNA with dimers (2 subunits)
sech unit has recognition helix, which increases specificity and stability
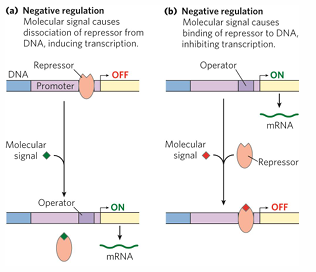
negative regulation
default for most bacterial is on, can be with inducible or repressible
repressor = protein blocks RNA poly from binding / moving along
operators = binding sites on DNA bind repressors near promoter
effectors = small molecule binds repressor and conformational change
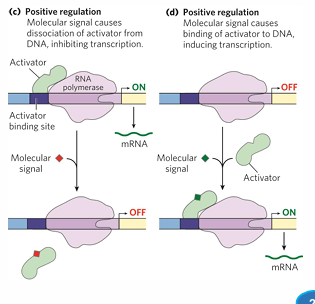
positive regulation
genes with weak promoters may need activators
activators = enhance RNA poly-promoter interaction
activator binding sites, effectors
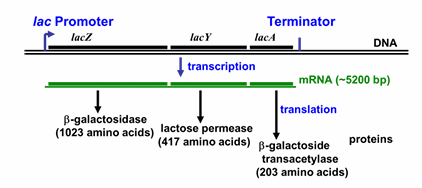
lac operon
3 genes do lactose metabolism,
in presence of lactose
lactose > galactose + glucose
lactose > allolactose
absence of glucose, adenylate cyclase makes cAMP
lac repressor
homotetramer, dimer of dimers
no lactose, repressor binds O1 and O2/O3
lactose > allolactose binds repressor to prevent it binding to DNA
CRP activator
lac operon has weak promoter
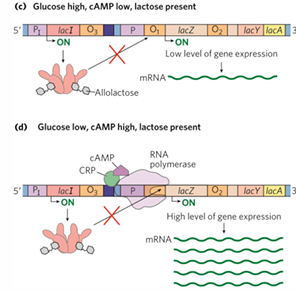
CRP
cAMP receptor protein;
contains binding sites for DNA and cAMP
homodimer, binds activator-binding site upstream promoter, interacts with RNA poly alpha
***assists RNA poly binding to lac promoter
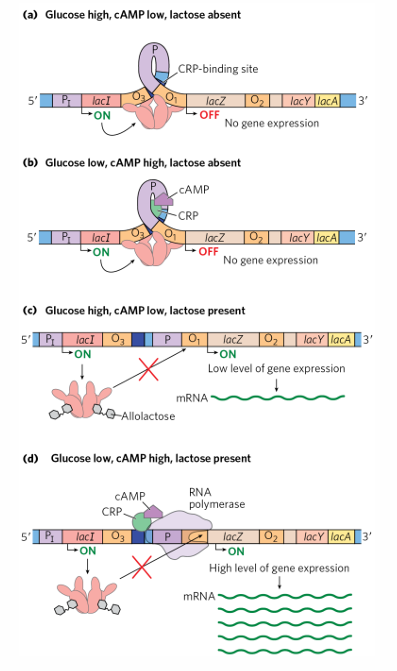
regulation of lac operon
negative =
lacI gene upstream of operon, encodes repressor
lac repressor binds operon in absence of allolactose
operator site at transcription start site
positive =
cAMP receptor protein binds DNA in presence of inducer cAMP
activator binding site located upstream of the lac promoter
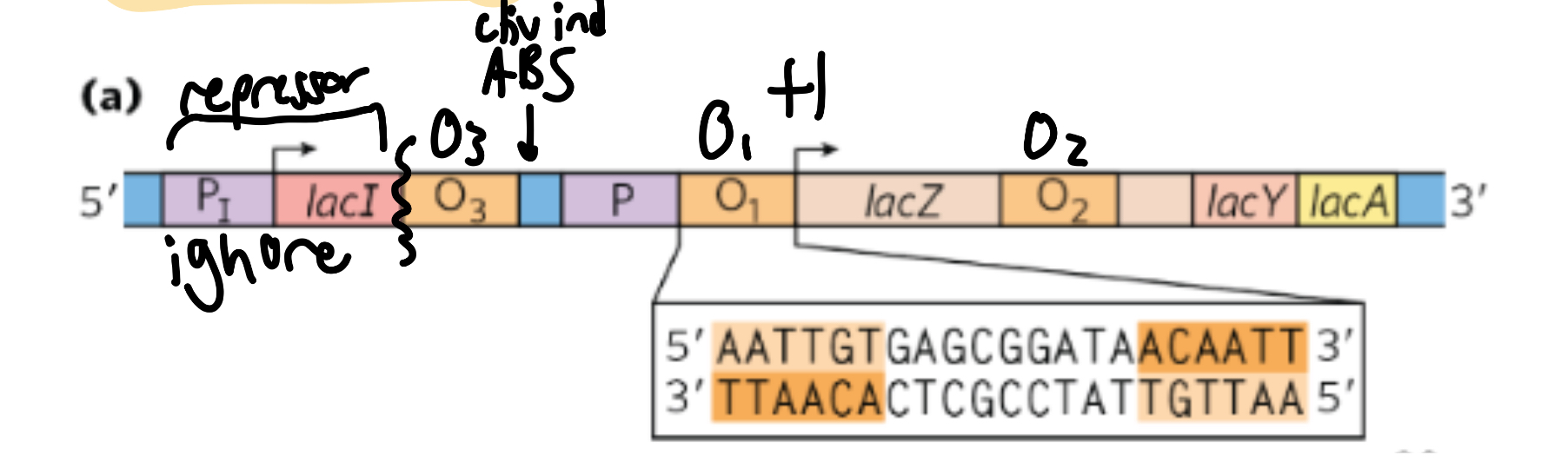
lac operon
visual
replication v transcription primers
RNA polymerases in transcription don’t use primers
who binds to the operator
repressors, NOT effectors (allolactose, which binds to proteins) or activators (which bind to enhancers or bind sites)