Biochemistry Exam 3 Study Guide
1/113
Earn XP
Description and Tags
I need to do well on tests can't let this situation happen again
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
114 Terms
metabolism
sum of chemical change converts nutrients into energy & chemically complex finished products of cellconverts
metabolic classification of organism based on carbon requirement & energy use
photoautotrophs (use CO2)
photoheterotrophs (use light)
chemoautotrophs (use organic & inorganic e- donor)
chemoheterotrophs (use inorganic carbon)
metabolic classification of organism based on O2 requirement
aerobes (+O2 as e- donor)
anaerobes (±O2 or -O2)
catabolism
degradation of complex molecules to simple ones
oxidative
exergonic: release energy
what pathways are catabolic?
glycolysis & citrate acid cycle
generalization about catabolic pathway
extract energy by oxidizing fuels
generate ATP
generate reduced electron carriers
converge to few intermediate
what two molecules are convergence points for sugar, fatty acid, and amino acid metabolism?
pyruvate & acetyl CoA
anabolism
biosynthesis of complex molecules from simple ones
reductive
endergonic: requires energy
what pathways are anabolic?
gluconeogenesis
generalization of anabolic pathway
reductive biosynthesis
common electron donor: NAHD
require energy from energy investment
in what form is carbon release when it is fully oxidized?
CO2, DOESN’T happens in glycolysis
what is oxidized during oxidative phosphorylation?
NADH & FADH2
what is reduced during oxidative phosphorylation?
O2
what is produced as product of electron transport from NADH & FADH2 to O2?
proton gradient
what is the proton gradient coupled with?
ATP synthesis
metabolic flux
controlled by allosteric control, covalent modification, substrate cycles, and generic control
high energy compounds
energy currency
why does transfer of a phosphoryl group from ATP have a highly negative delta G?
resonance stabilization is greater in product
reduced electrostatic repulsion
hydrolysis products are better solvated
substrate level phosphorylation
high energy molecule that are power to transfer of phosphoryl group from a phosphorylated subtract to ADP
how to regenerate ATP from ADP?
subtract level phosphorylation
oxidative phosphorylation
photophosphorylation
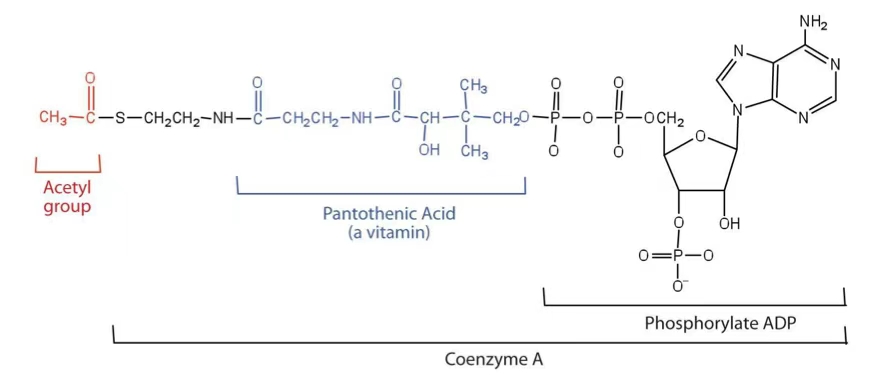
acetyl CoA
high energy compound
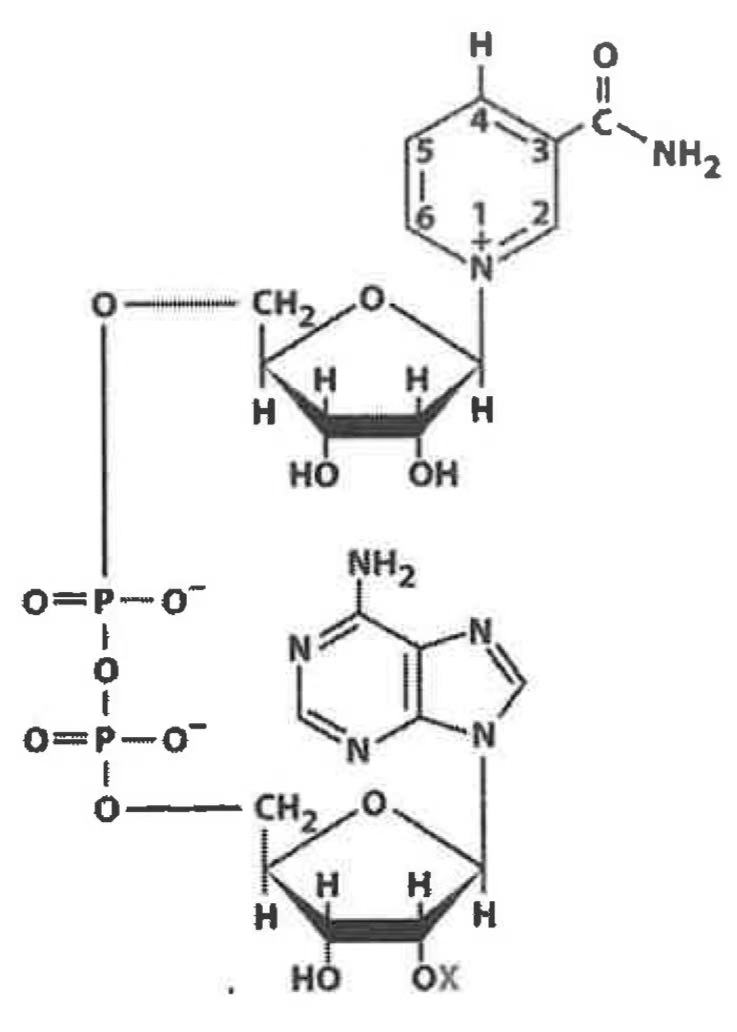
NAD+
only accepts/donates 2 electrons at a time in the form of hydride (1 proton/2 electron)
redox of alcohols & carbonyls
co-substrate for many dehydrogenase
derived from nictin B3
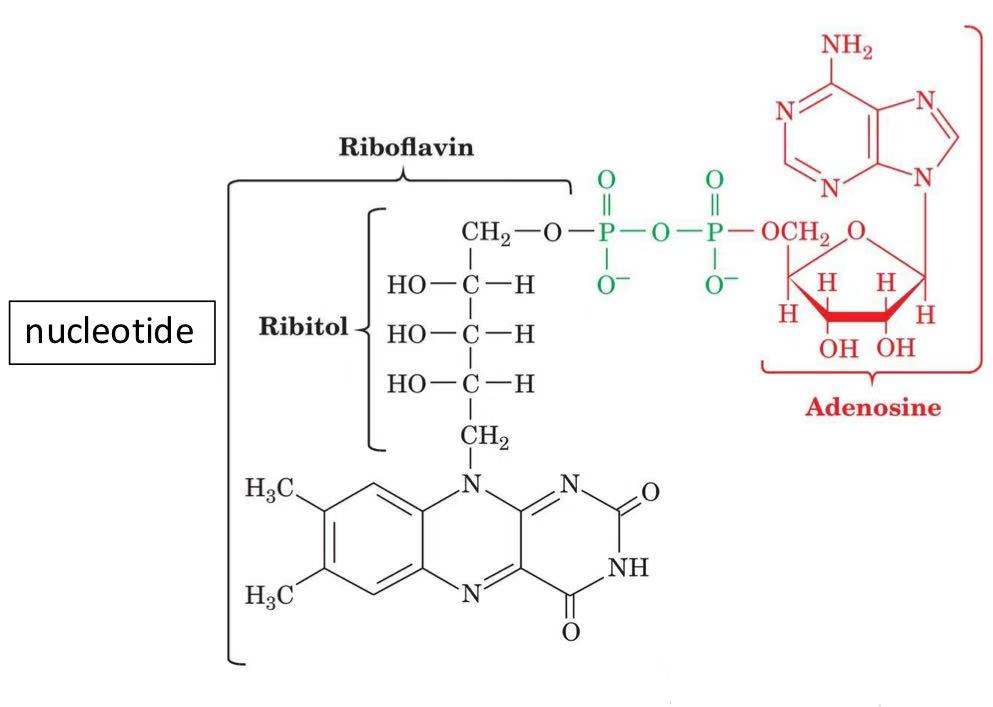
FAD
accepts 1/2 electrons at a time in the form of 1 proton/1 electron
redox of alkanes
prosthetic group
derived from riboflavin B2
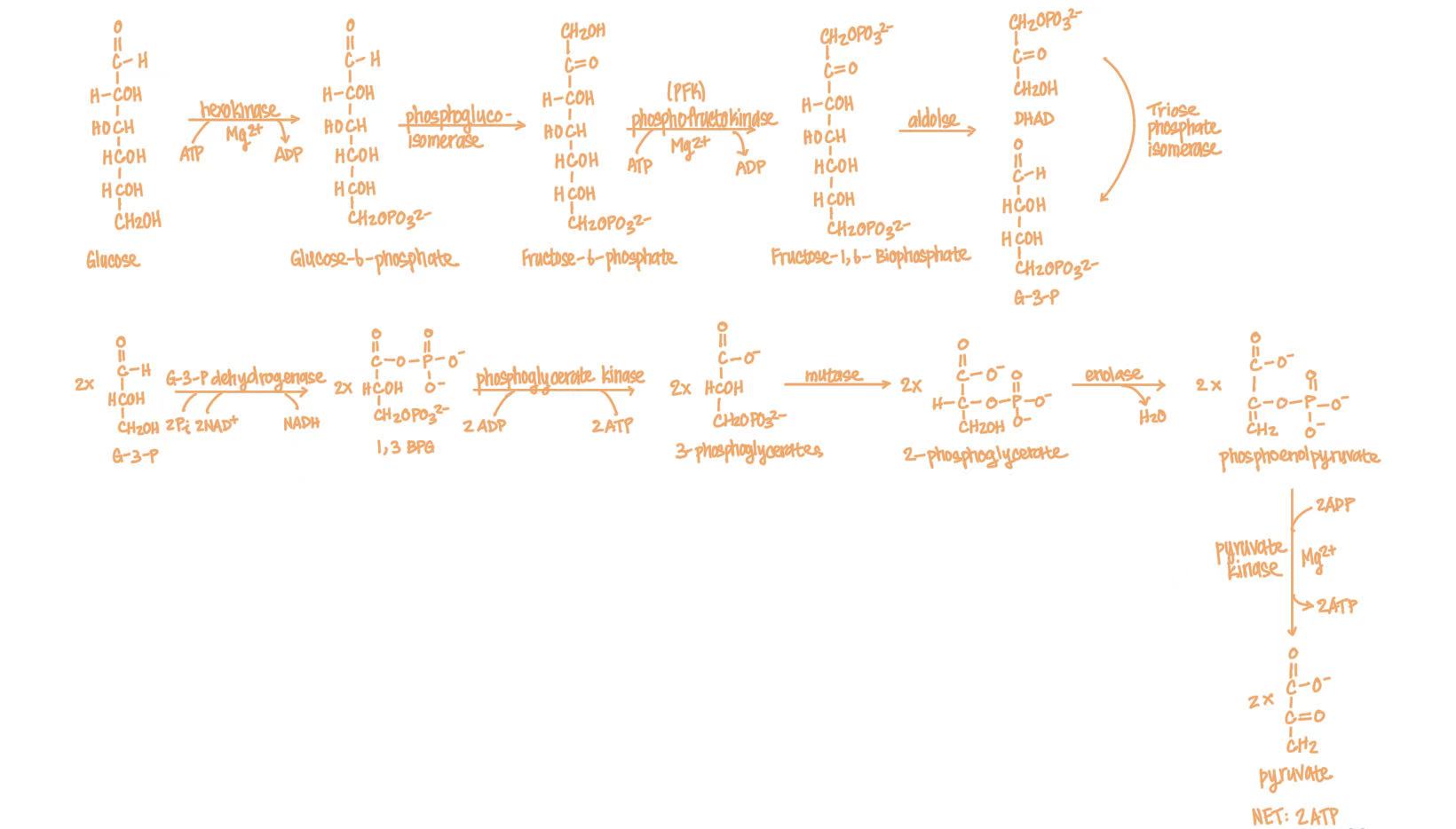
glycolysis
The metabolic process where glucose is converted into pyruvate, producing small amounts of ATP and NADH.
glycolysis process
glucose (hexokinase, Mg2+, ATP→ADP) glucose-6-phosphate
glucose-6-phosphate (phosphoglucoisomerase) fructose-6-phosphate
fructose-6-phosphate (phosphofructokinase, Mg2+, ATP→ADP) fructose-1,6-bisphosphate
fructose-1,6-bisphosphate (aldolase) dihydroxyacetone phosphate & gluceraldehyde-3-phosphate
dihydroxyacetone phosphate (triose phosphate isomerase) gluceraldehyde-3-phosphate
2x gluceraldehyde-3-phosphate (gluceraldehyde-3-phosphate dehydrogenase, NAD+→NADH) 1,3-bisphosphoglycerate
2x 1,3-bisphosphoglycerate (phosphoglycerate kinase, Mg2+, ADP→ATP) 3-phosphoglycerate
2x 3-phosphoglycerate (phosphoglycerate mutase) 2-phosphoglycerate
2x 2-phosphoglycerate (enolase, Mg2+, H2O) phosphoenolpyruvate
2x phosphoenolpyruvate (pyruvate kinase, Mg2+, K+, ADP→ATP) pyruvate
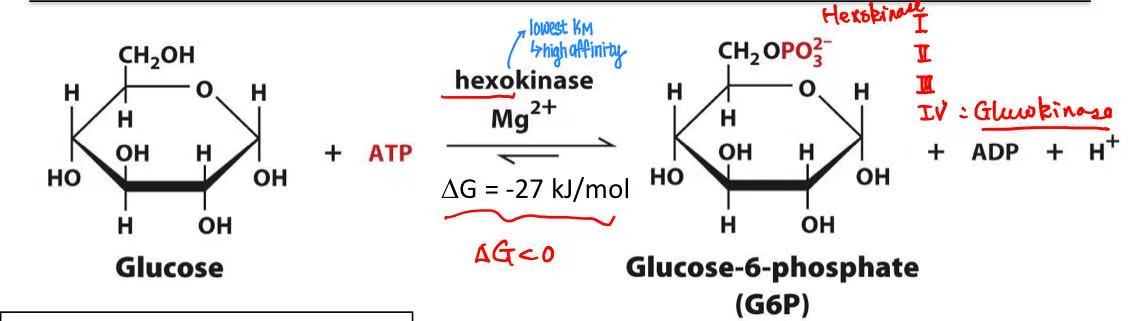
glucose + ATP → glucose-6-phosphate
hexokinase
Mg2+
hexokinase mechanism
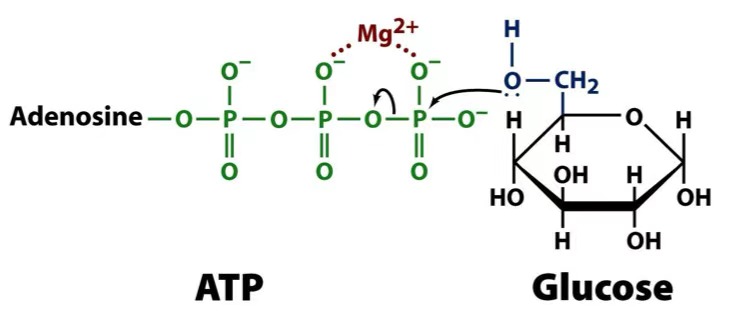
hexokinase can also phosphorylate
nonspecific: glucose, mannose, fructose
what does Mg2+ do?
shield negative charges of phosphate group
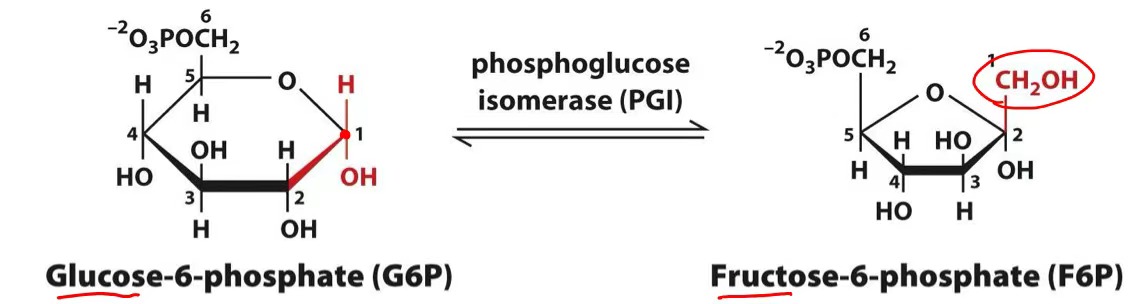
glucose-6-phosphate → fructose-6-phosphate
phosphoglucoisomerase
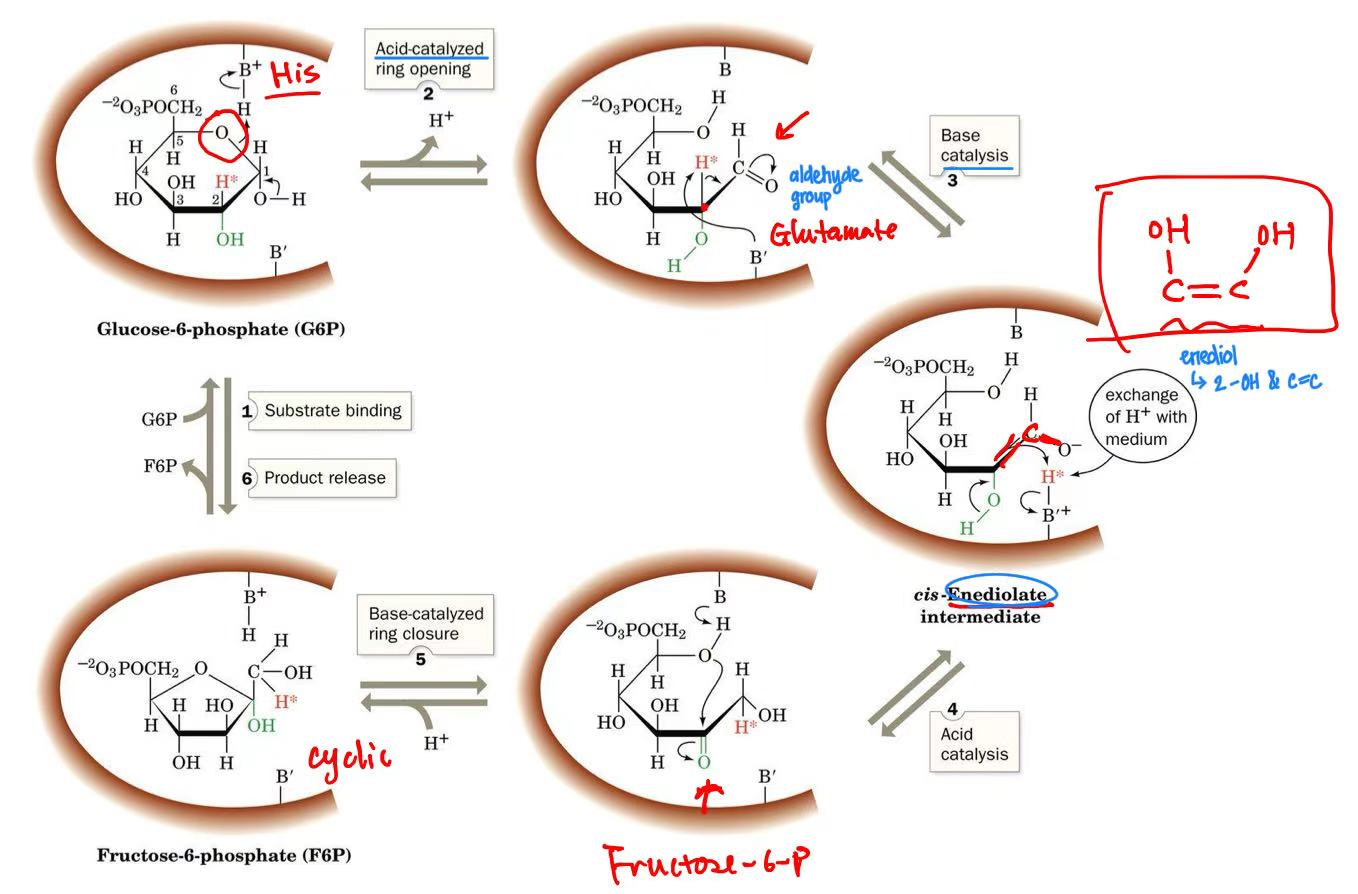
phosphoglucoisomerase mechanism
acid catalysis → open ring
intermediate: enediol (2 -OH & 1 C=C)
basic catalysis → close ring
fructose-6-phosphate + ATP → fructose-1,6-bisphosphate
phosphofructokinase
Mg2+
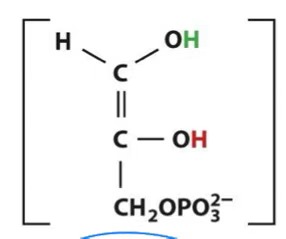
fructose-1,6-bisphosphate → glyceraldehyde-6-phosphate + dihydroxyacetone phosphate
aldolase
aldolase mechanism
schiff base
covalent catalysis
aldol cleavage
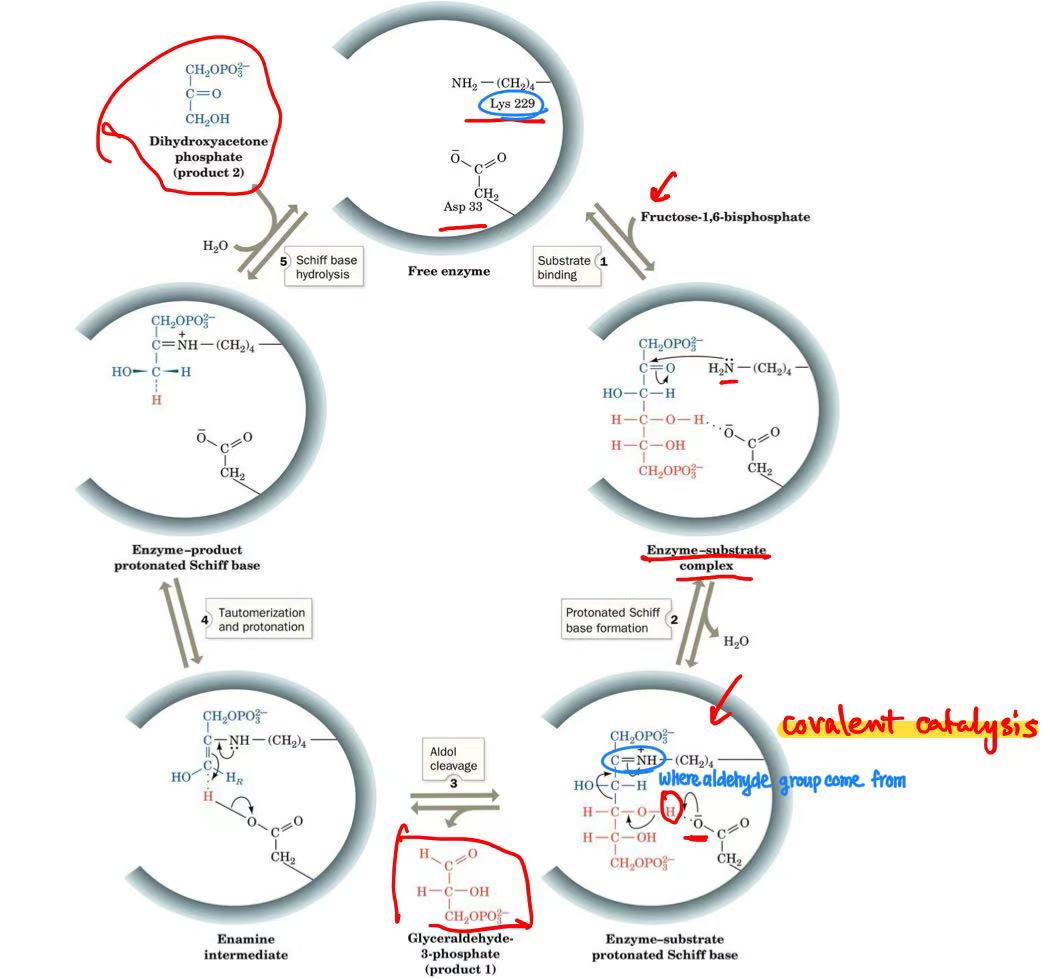
dihydroxyacetone phosphate→glyceraldehyde-6-phosphate
triose phosphate isomerase
triose phosphate isomerase intermediate
enediol
glyceraldehyde-3-phosphate→1,3 bisphosphoglycerate+NADH
glyceraldehyde-3-phosphate dehydrogenase
glyceraldehyde-3-phosphate dehydrogenase mechanism
intermediate: thiohemiacetal intermediate & acyl thioester
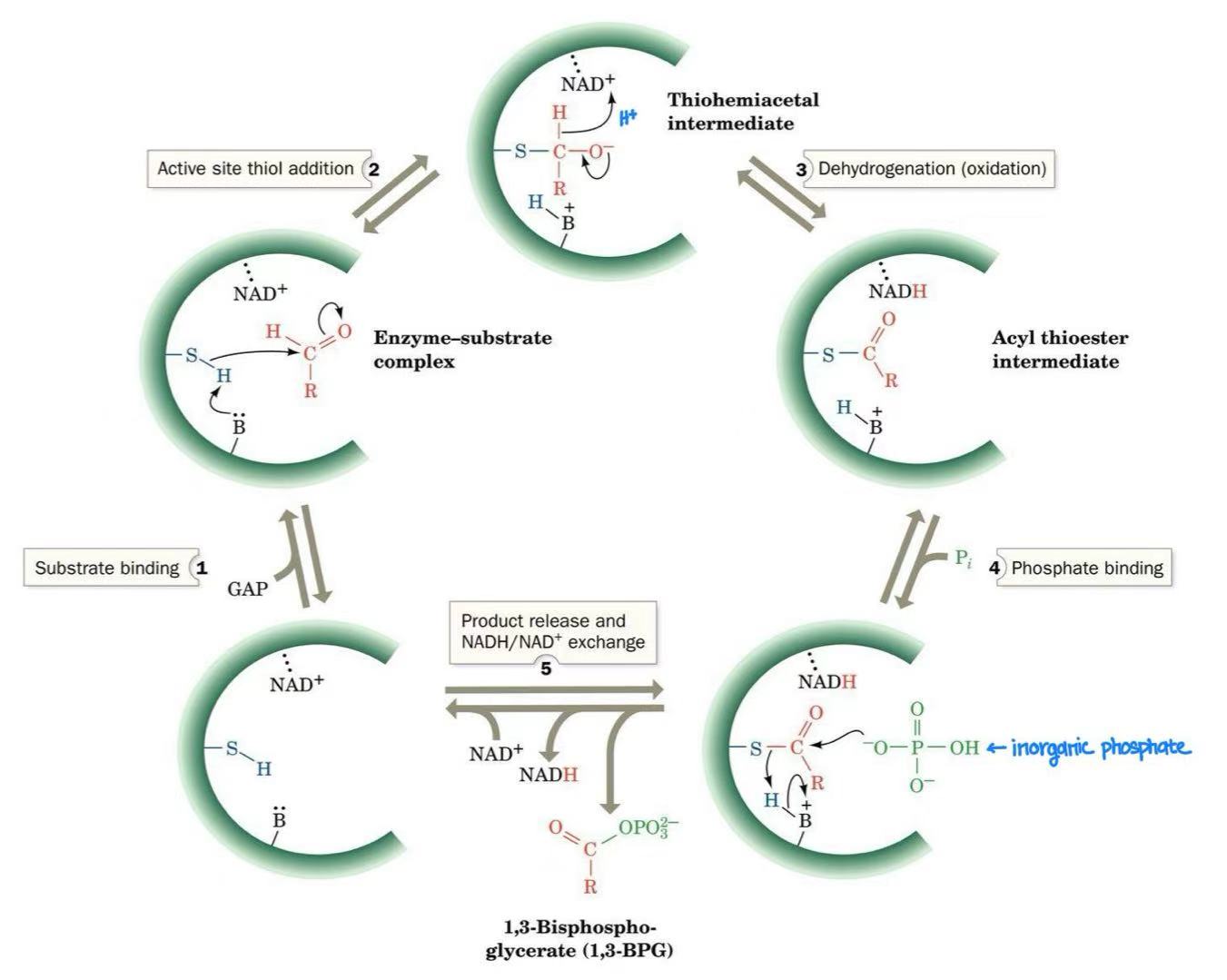
1,3-bisphosphoglycerate → 3-phosphoglycerate + ATP
phosphoglycerate kinase
Mg2+
3-phosphoglycerate → 2-phosphoglycerate
mutase
mutase mechanism
phosphorylated histidine
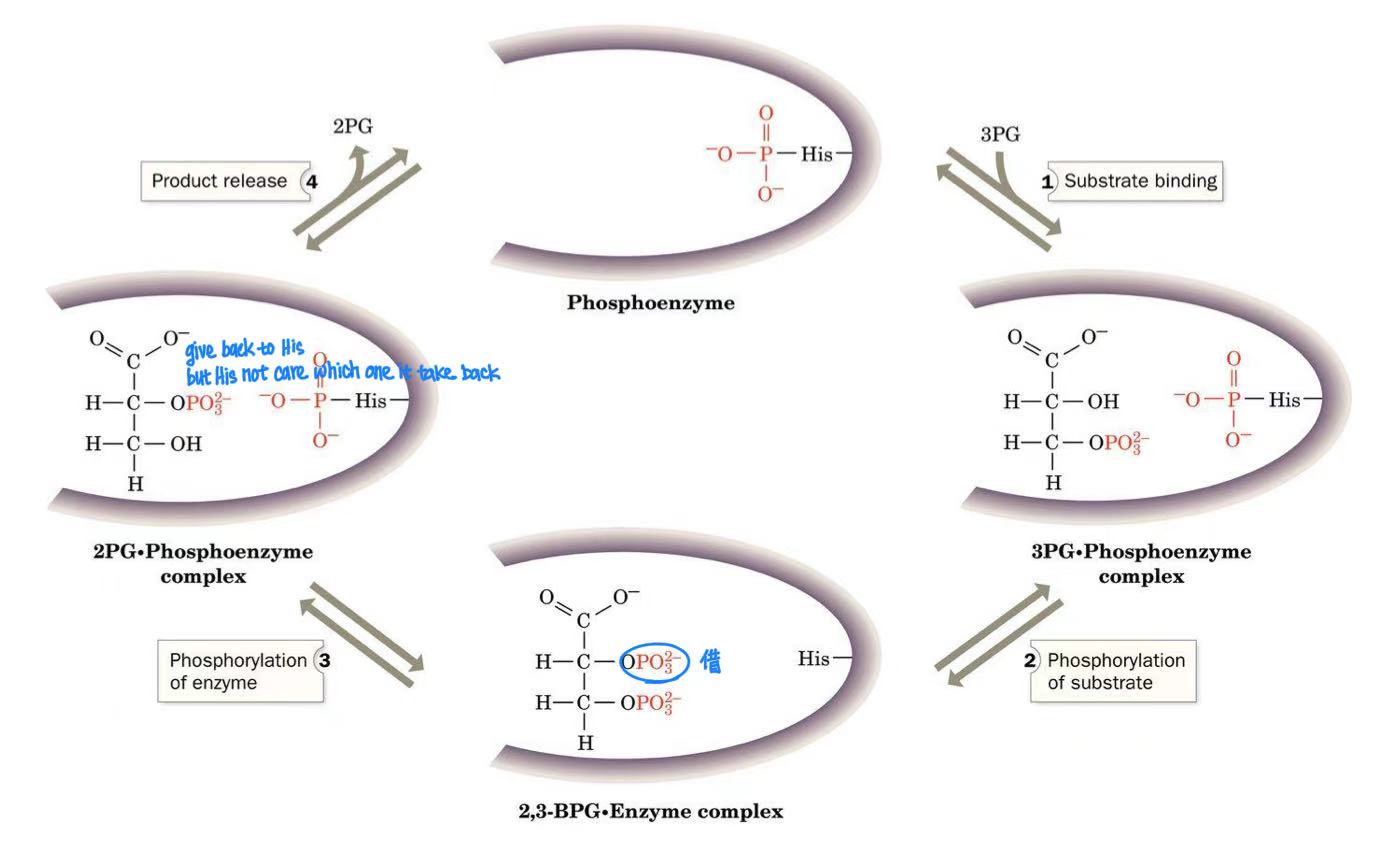
2-phosphoglycerate → phosphoenolpyruvate
enolase
phosphoenolpyruvate → pyruvate + ATP
pyruvate kinase
pyruvate + NADH → lactate
homolactic fermentation: lactate dehydrogenase
reduction rxn
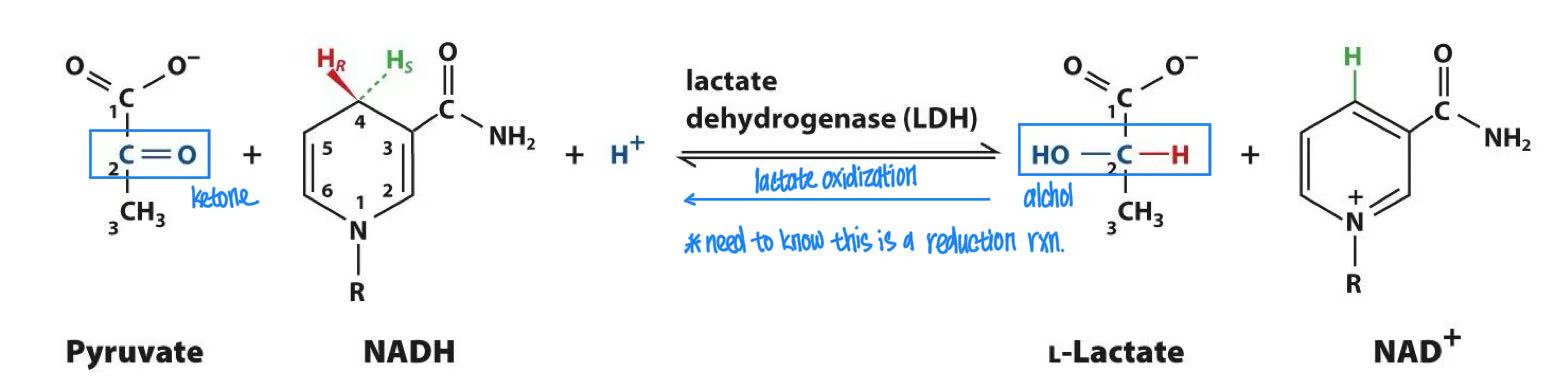
glycolysis regulation
step 1: hexokinase
step 3: phosphofructokinase
step 10: pyruvate kinase
phosphofructokinase regulation
fructose-6-phosphate → fructose-1,6-bisphosphate
(-) ATP, citrate, phosphoenolpyruvate
(+) AMP/ADP, fructose-2,6-bisphosphate
pyruvate→acetyl-CoA process
happens in mitochondria matrix
pyruvate + thiamine pyrophosphate (pyruvate dehydrogenase) hydroxyethyl-thiamine pyrophosphate + CO2
acetyl group + lipoamide (dihydrolipoyl transacetylase) acetyl-dihydrolipoamide
acetyl-dihydrolipoamide + CoA-SH (dihydrolipoyl transacetylation) acetyl-CoA + dihydrolipoamide
dihydrolipoamide (oxidized dihydrolipoyl dehydrogenase) reduced dihydrolipoyl dehydrogenase + lipoamide
dihydrolipoyl dehydrogenase (NAD+→NADH+H+) oxidized dihyrolipoyl dehydrogenase
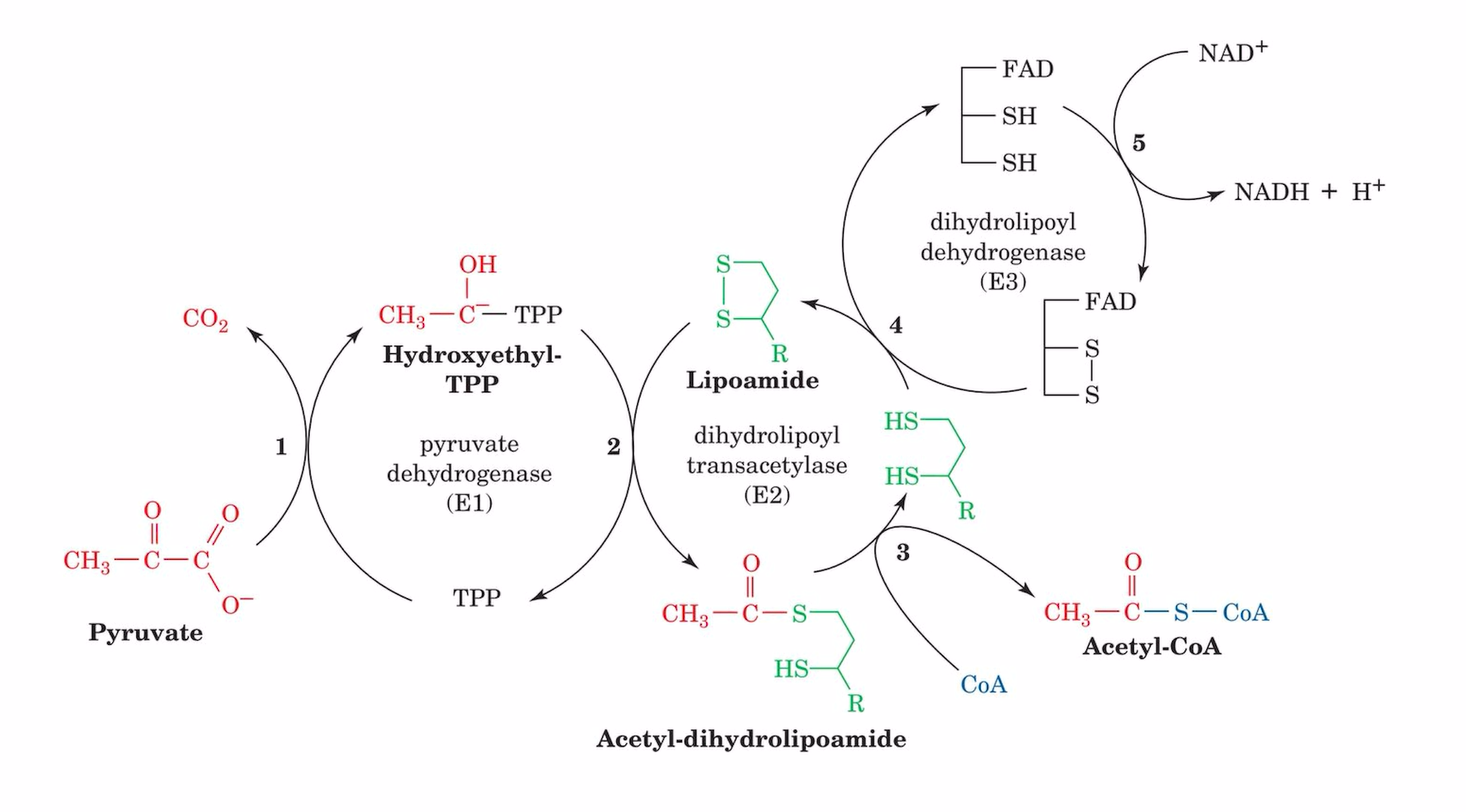
E1
pyruvate dehydrogenase: an enzyme complex that converts pyruvate into acetyl-CoA, linking glycolysis and the citric acid cycle.
TPP = thiamine pyrophosphate
E2
dihydrolipoyl transacetylase
lipoic acid & CoA
E3
dihydrolipoyl dehydrogenase
FAD & NAD+
benefits of multienzyme complex
increase reaction rate
reduced competing reaction
coordinate control
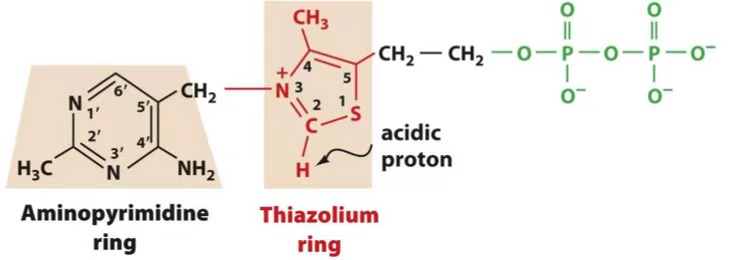
TPP
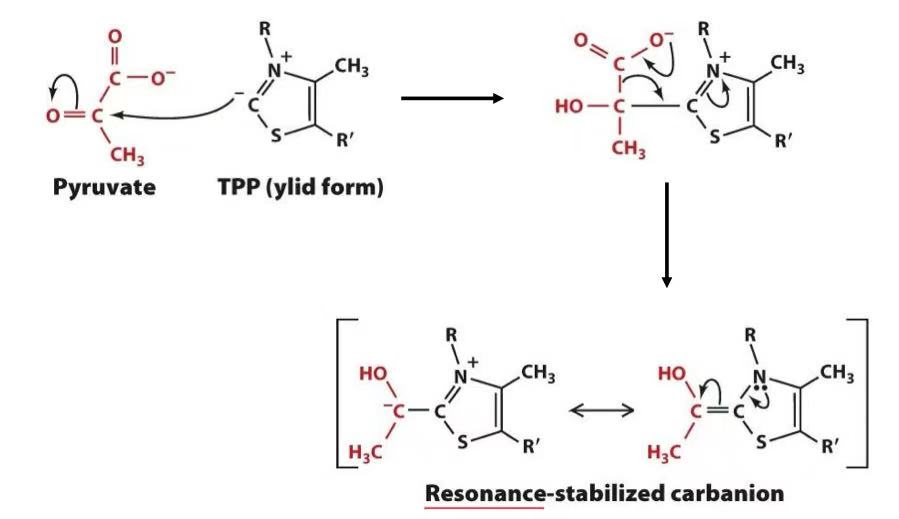
decarboxylation of pyruvate (losing CO2)
acetylation of lipoamide
acetyl oxidized reacts with disulfide & disulfide reduced
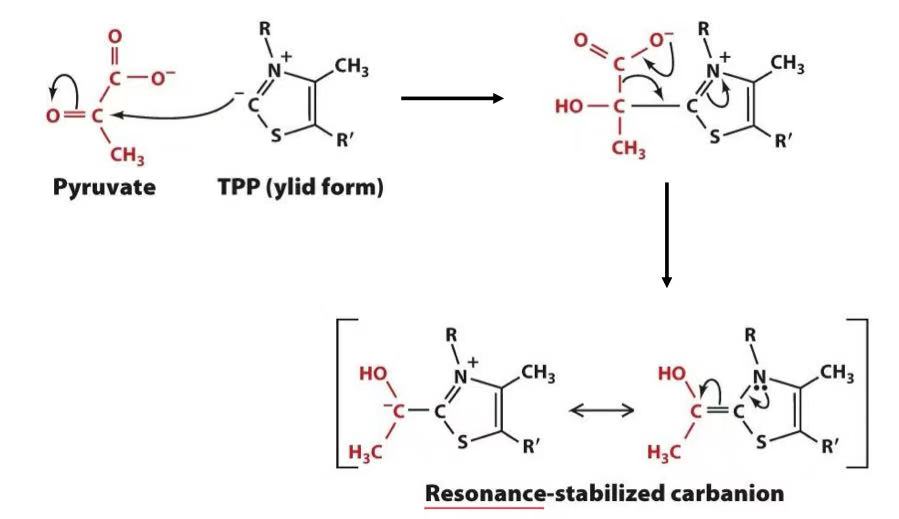
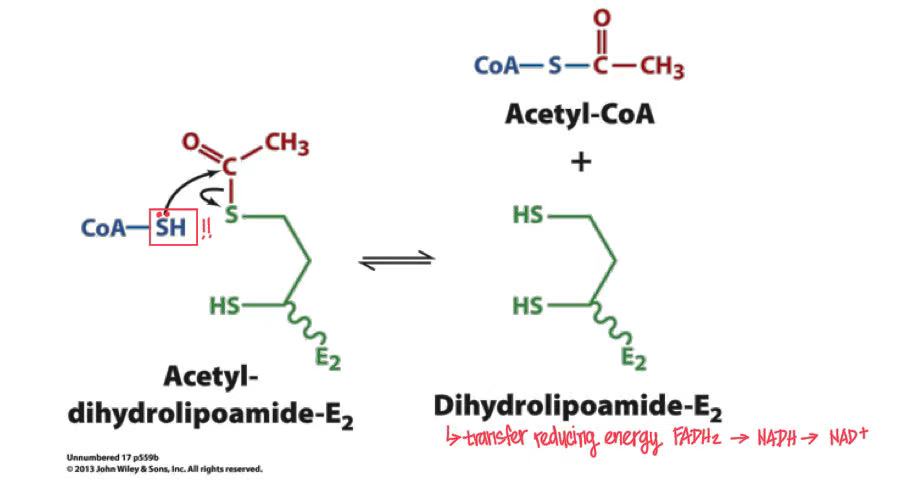
acetylation of CoA
CoA reacts with acetyl-lipoamide → acetyl-CoA goes to citrate acid cycle & dihydrolipoamide release
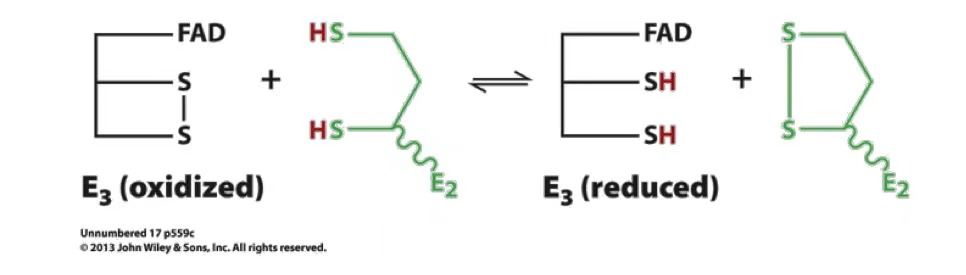
regeneration of lipoamide
FAD cofactor accepts 1 electron at a time in form of H
NAD+ cofactor accepts 2 electron and H+
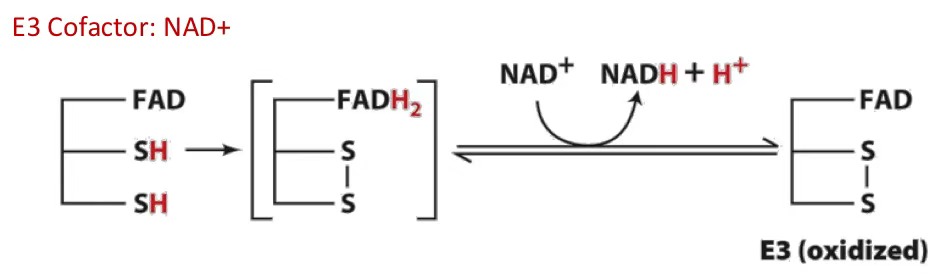
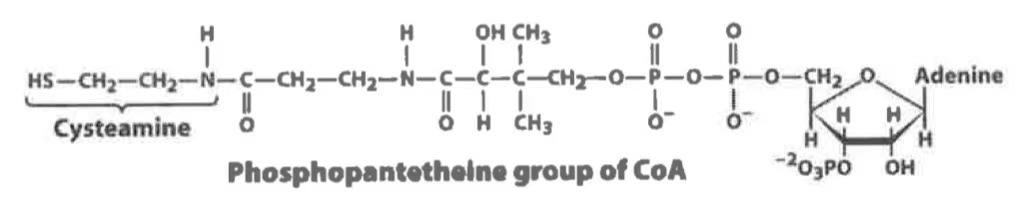
coenzyme A (CoA)
A coenzyme that carries acyl groups in the form of acyl-CoA, vital for various metabolic pathways.
what is pyruvate dehydrogenase highly regulated by?
allosteric inhibition
NADH
Acetyl-CoA
covalent modification (phosphorylation/dephosphorylation)
activated & dephosphorylate by insulin & Ca2+
inhibited & phosphorlated by acetyl-CoA & NADH
permanent inhibition by arsenic
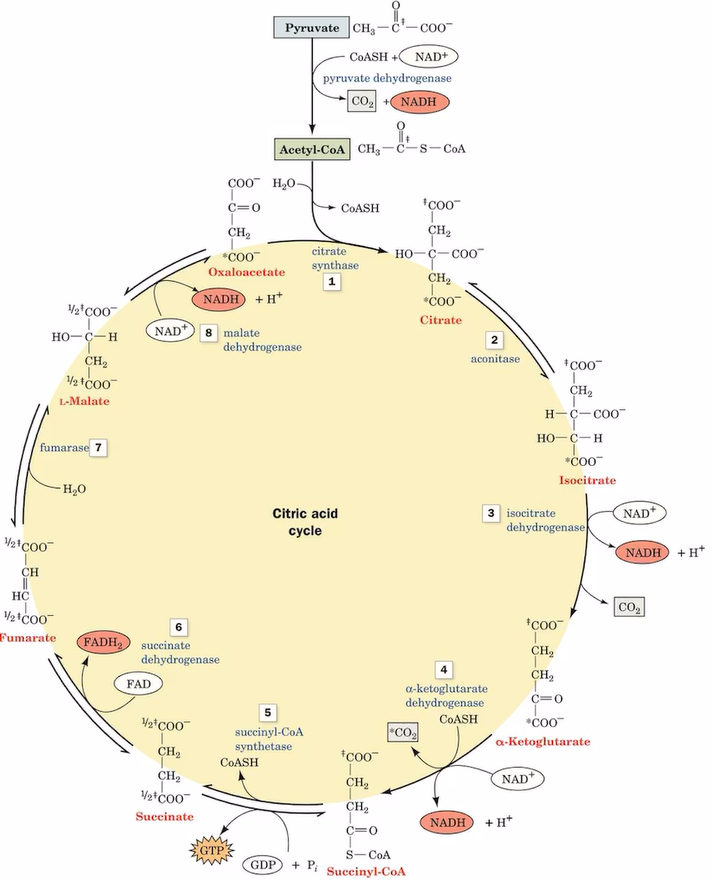
citrate acid cycle
a series of chemical reactions used by all aerobic organisms to release stored energy through the oxidation of acetyl-CoA.
take place in mitochondria
generation of citric acid cycle
catabolism
carbon atom oxidized to CO2
electron transferred to electron carries
electron ultimately transferred to O2
citrate acid cycle process
acetyl-CoA + oxaloacetate (citrate synthase, H2O) citrate + CoA-SH
citrate (aconitase) isocitrate
isocitrate (isocitrate dehydrogenase, NAD+→NADH, CO2) alpha-ketoglutarate
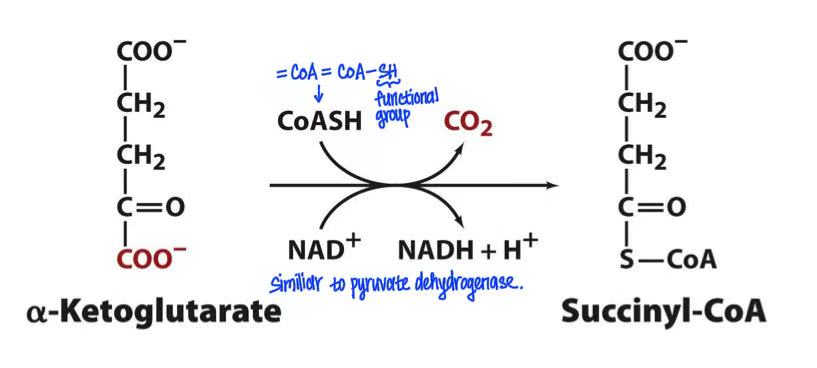
alpha-ketoglutarate (alpha-ketoglutarate dehydrogenase, NAD+→NADH, CoA-SH→CO2) succinyl-CoA
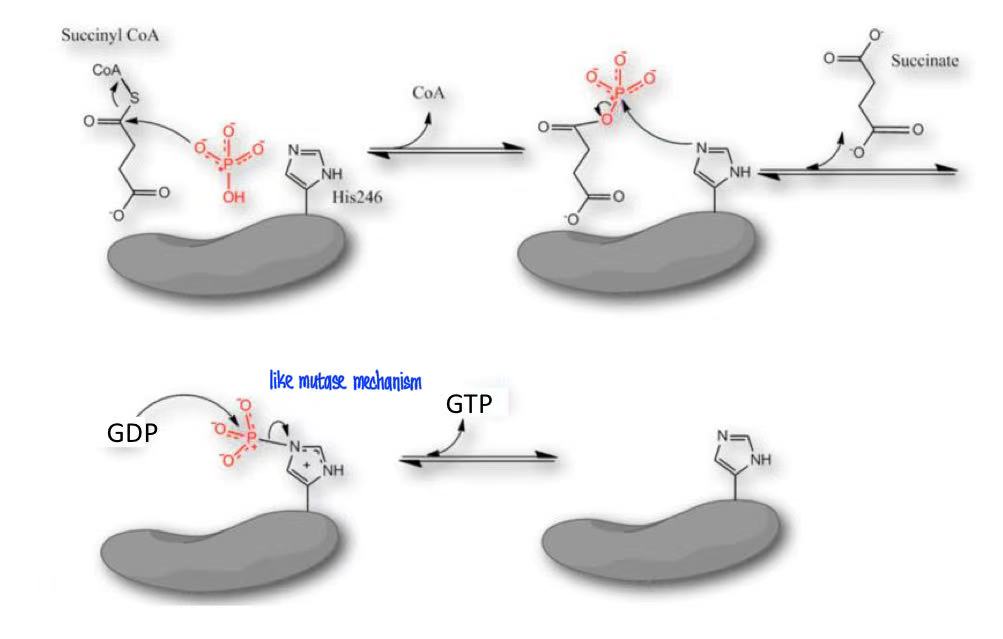
succinyl-CoA (succinyl-CoA synthetase, CoASH, ADP→ATP) succinate
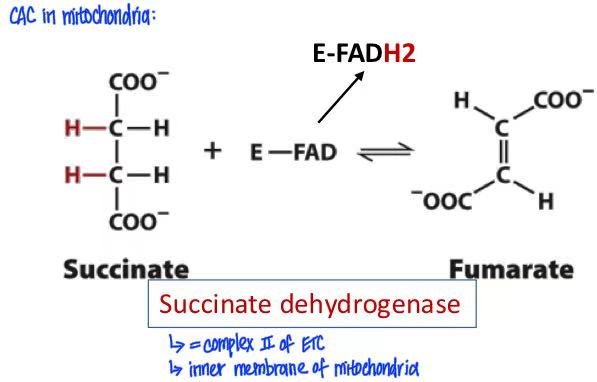
succinate (succinate dehydrogenase, FAD→FADH2) fumarate
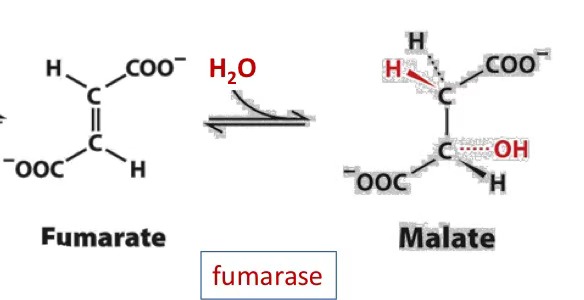
fumarate (H2O, fumarase) malate
malate (malate dehydrogenase, NAD+→NADH) oxaloacetate
what are the regulators in citrate acid cycle?
acetyl-CoA, oxaloacetate, NADH, (ATP)
distributed through 3 steps (delta G<0):
citrate synthase (acetyl-CoA + oxaloacetate → citrate)
isocitrate dehydrogenase (isocitrate → alpha-ketoglutarate) uses NAD+→NADH
alpha-ketoglutarate dehydrogenase (alpha-ketoglutarate → succinyl-CoA) uses NAD+→NADH
supply & demand: substrate availability, product inhibition, competitive feedback inhibition
oxaloacetate + acetyl-CoA → citrate + CoA
citrate synthase
citrate synthase mechansim
rate limiting formation of acetyl-CoA enolate by Asp & hydrogen bond stabilized by His
His basic catalysis → acetyl-CoA nucleophilic attack oxaloacetate
citryl-CoA hydrolysis
intermediate: citryl-CoA
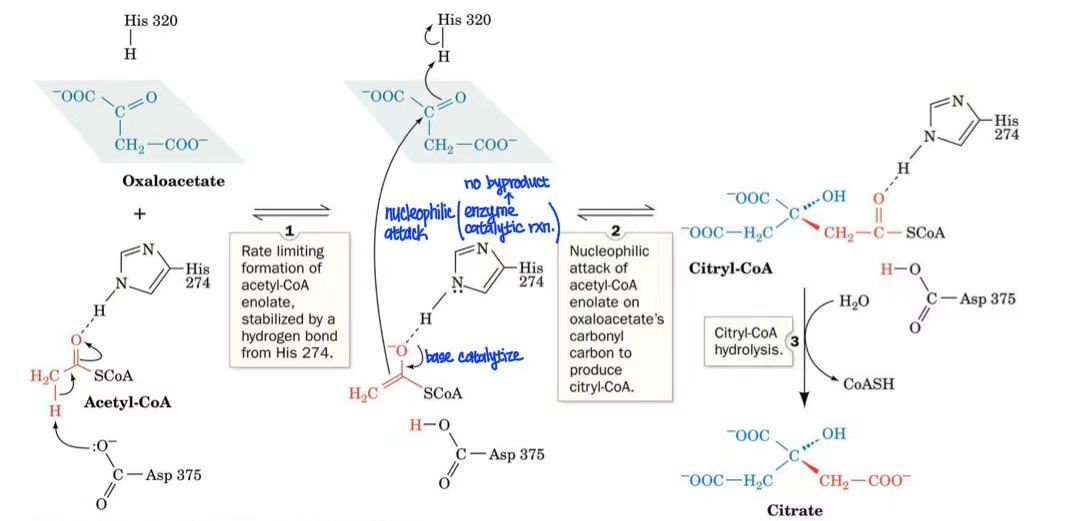
what regulates citrates synthase?
(-) NADH, citrate, succinyl-CoA
citrate → isocitrate
aconitase
isocitrate → alpha-ketoglutarate + NADH
isocitrate dehydrogenase
isocitrate dehydrogenase
decarboxylation of isocitrate
oxidation of alcohol → ketone
decarboxylate
tautomerization
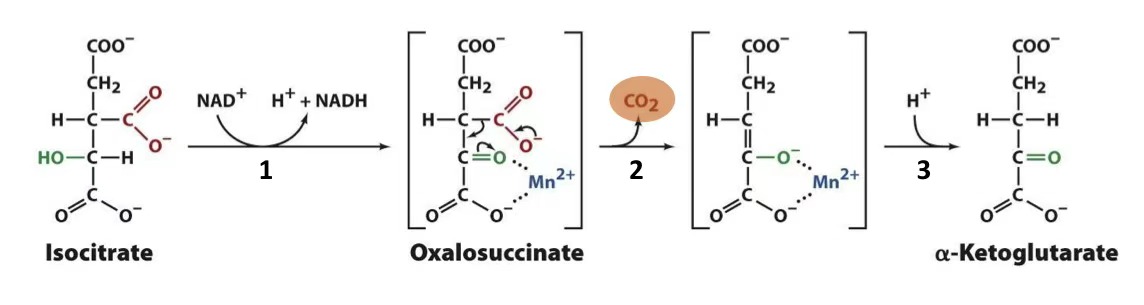
what regulates isocitrate dehydrogenase?
(+) Ca2+, ADP
(-) ATP, NADH
alpha-ketoglutarate → succinyl-CoA + NADH + CO2
alpha-ketoglutarate dehydrogenase
resemble pyruvate dehydrogenase
TPP, lipoamide, CoASH
what regulates alpha-ketoglutarate?
(+) Ca2+, AMP
(-) succinyl-CoA, NADH
succinyl-CoA → succinate + GTP + CoA
succinyl-CoA synthetase (thiokinase)
succinyl-CoA synthetase
subtract-level phosphorylation
phosphorylated His
succinate → fumarate + FADH2
succinate dehydrogenase
fumarate → malate
fumarase
malate → oxaloacetate + NADH
malate dehydrogenase
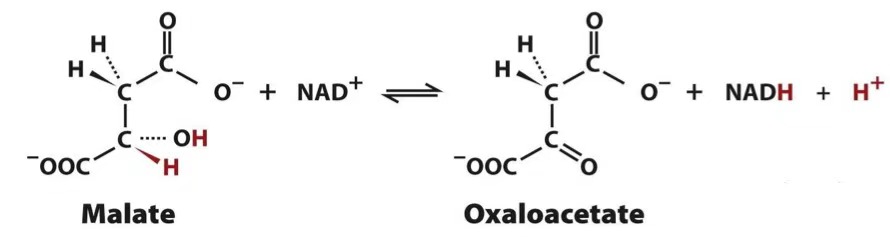
malate-aspartate shuttle
reduces oxaloacetate to malate which crosses into matrix & reduces NAD+ → NADH
start as NADH in cytosol & goes into mitochondria as NADH
glycerophosphate shuttle
supplies electron directly to ETC: dihydroxyacetone phosphate → 3-phosphoglycerol using NADH, this donates electron to FAD already embedded in the membrane
start as NADH in the cytosol but become FADH in mitochondira
electron transport chain
a series of protein complexes in the inner mitochondrial membrane that transfer electrons and pump protons to create a gradient.
electron transport chain process
NADH→complex I→coenzymeQ→complex III→cytochrome c→complex IV→oxygen
succinate→complex II (succinate dehydrogenase)→coenzymeQ→complex III→cytochrome c→complex IV→oxygen
glycerophosphate shuttle (NADH from cytosol→FADH2 in mitochondria)→coenzymeQ→complex III→cytochrome c→complex IV→oxygen
fatty acyl-CoA dehydrogenase (FAD→FADH2)→coenzymeQ→complex III→cytochrome c→complex IV→oxygen
generation of electron transport chain and oxidative phosphorylation
NADH & FADH2 deoxidized → go back to glycolysis & CAC cycle
electron ultimately reduced O2 → H2O
electron transfer & expulsion of H+ produces proton gradient driving ATP synthesis
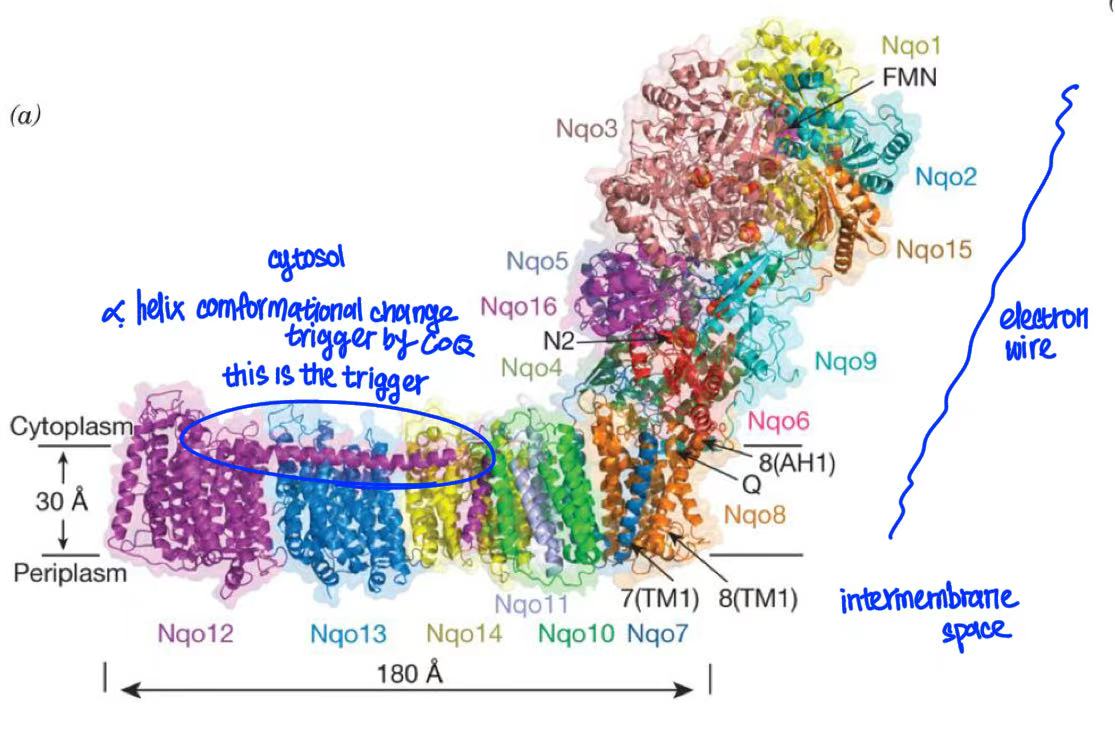
complex I
oxidation of NADH by CoQ
NADH donates 2 electrons to FMN → electron passes through Fe-S cluster → to CoQ
when the electrons move from one redox center to another, they create a redox wire & generate energy
every pair of electrons results in 4 protons transferred from the matrix to the intermembrane space
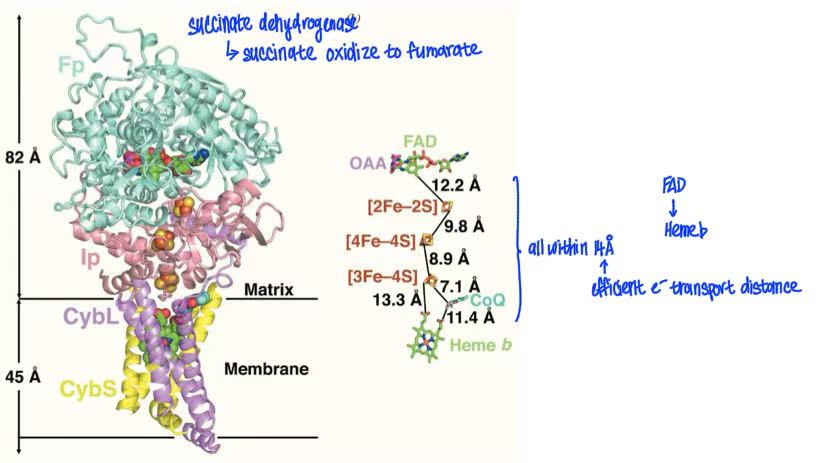
complex II = succinate dehydrogenase
oxidation of FADH2 by CoQ
FADH2 donates 2 electrons to complex II → pass through Fe-S cultures → to CoQ
complex III
oxidation of CoQ to Cyt C
CoQ from complex I and complex II donates electron to Cyt C in complex III → electrons travel through hemes & Fe-S clusters
CoQ can donate 2 electrons but Cyc C only accepts 1 electron so 2 electrons have different paths of Q cycle
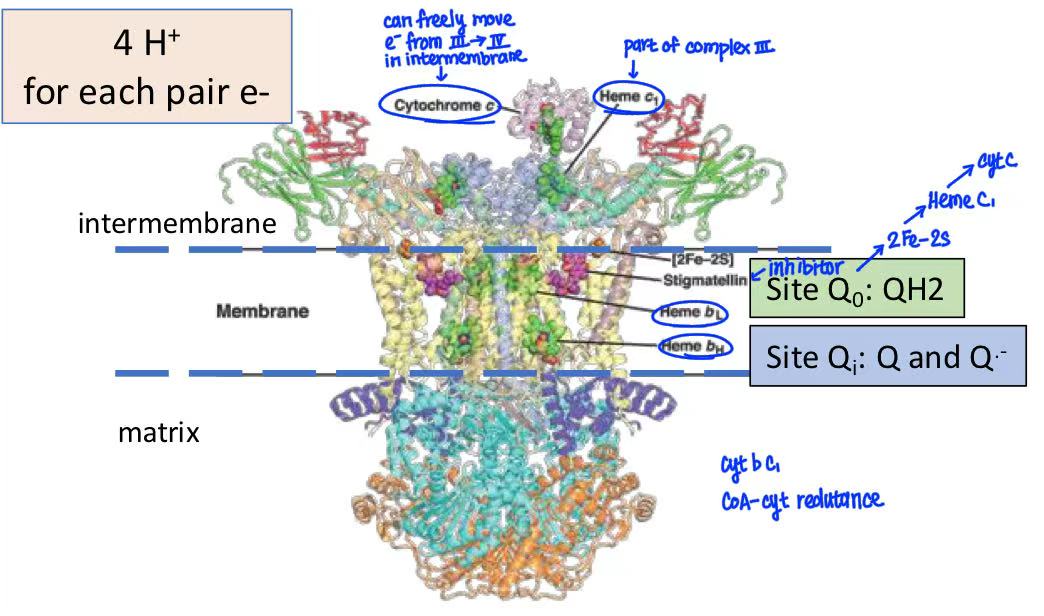
Q cycle
complex IV
oxidation of CytC by O2
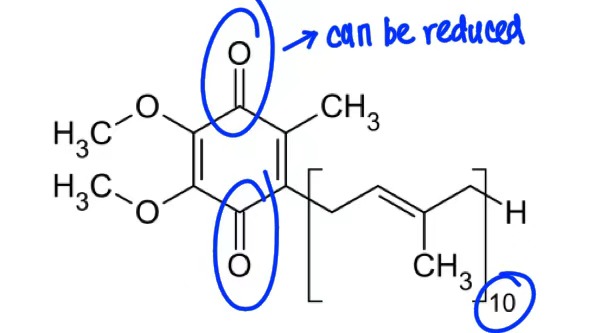
Coenzyme Q
transport electrons from complex I & II → III
affiliated with I, II, III
accept 1 or 2 electron
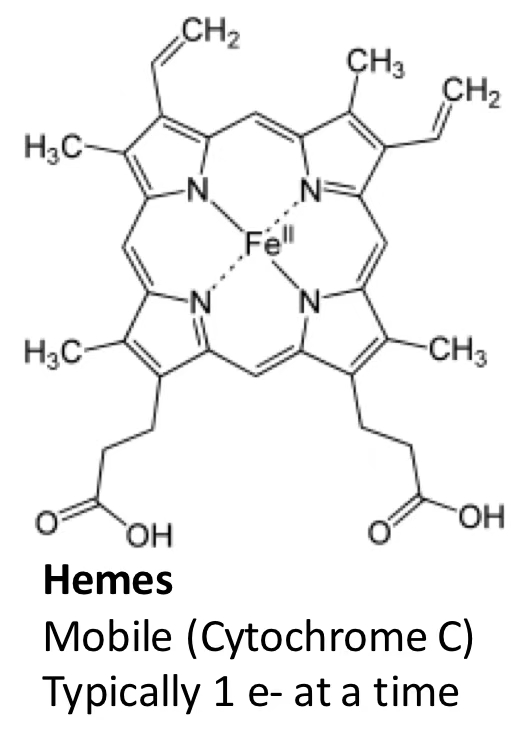
Cytochrome C
transport electron from complex III → IV
accept 1 electron
flavin mononucleotide (FMN)
affiliate with complex I
accept 1 or 2 electrons
Fe-S cluster
affiliated with I, II, III
accept 1 electron
what is the final electron acceptor?
O2
oxidative phosphorylation
synthesis ATP
gluconeogenesis
The synthesis of glucose from non-carbohydrate precursors, primarily occurring in the liver.
sources of glucose through gluconeogenesis
glycogen (glucose-6-phospate can be store at glycogen)
all amino acid except lysine & leucine
all citrate acid cycle intermediates
glycerol from triacylglycerols (NOT fatty acid)
is pyruvate → acetyl-CoA reversible?
NO
gluconeogenesis process
pyruvate (pyruvate carboxylase, ATP+CO2→ADP+Pi) oxaloacetate
oxaloacetate (phosphoenolpyruvate carboxykinase, GTP→GDP+CO2) phosphoenolpyruvate
phosphoenolpyruvate (enolase) 2-phosphoglycerate
2-phosphoglycerate (mutase) 3-phosphoglycerate
3-phosphoglycerate (phosphoglycerate kinase, ATP→ADP) 1,3-bisphosphoglycerate
1,3-bisphosphoglycerate (glyceraldehyde-3-phosphate dehydrogenase, NAD+Pi→NADH+H+) glyceraldehyde-3-phosphate
glyceraldehyde-3-phosphate (triose phosphate isomerase) dihydroxyacetone phosphate
glyceraldehyde-3-phosphate + dihydroxyacetone phosphate (aldose) fructose-1,3-bisphosphate
fructose-1,3-bisphosphate (fructose bisphosphatase, H2O→Pi) fructose-6-phosphate
fructose-6-phosphate (phosphoglucose isomerase) glucose-6-phosphate
glucose-6-phosphate (glucose-6-phosphatase, H2O→Pi) glucose
requirement of pyruvate to phosphoenolpyruvate
2 reactions
pyruvate →oxaloacetate
Mt.
oxaloacetate →phosphoenolpyruvate
Mt/cytosol depends on the organism
2 enzymes
pyruvate carboxylase: with HCO3-+ATP→ADP+Pi
require coenzyme biotin
phosphoenolpyruvate carboxykinase (PEPCK): GTP→GDP+Pi
which enzyme require biotin?
pyruvate carboxylase