MBS 344: Exam 3
1/107
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
108 Terms
What are some differences between constitutive and regulated genes? Provide some examples of both.
constitutive→ always expressed at same rate (aka housekeeping genes)
ex. rRNA or GTF genes
regulated→ genes expressed in response to change (depend on needs of the cell)
ex. metabolism genes like lactose genes
What is differential gene expression? Is it observed in unicellular organisms, multicellular organisms, or both?
different cell types express different genes→ always diff cell types to perform diff functions
ONLY multicellular orgs bc unicellular need to be able to perform all the functions of multi-cellular in one cell
Describe three different strategies used by cells to regulate expression of genes.
transcriptional control: regulate activity of RNA polymerase (control amount of RNA)
translational control: regulate activity of ribosome and RNA stability (control amount of protein)
post-translational control: regulate protein activity using chemical mods on aa (control protein function)
Describe the differences between general transcription factors and regulatory transcription factors.
GTF: binds promoter and recruit RNA pol to template strand (require all 5)
RTF (activator/repressor): bind regulatory sequences and regulate rate of transcription by influencing ability of RNA pol to transcribe specific gene→ allows differential gene expression
Activators vs repressors. What are they? How do they regulate gene expression? How are they the same? How are they different?
both: RTF (bind reg sequences on DNA) and always expressed
activators bind enhancers→ increase rate of tranxn
promote RNA pol phosphorylation
accelerate TFIID binding DNA
repressors bind silencers→ decrease rate tranxn
masks sequence
hinder binding
block RNA pol phosphorylation
Why is DNA looping useful in gene regulation? How is DNA looping achieved?
mechanism used to bring activators closer to promoter
architectural regulators→ proteins that bind specific sequences in bt regulatory sequences and promoter and bend DNA
What are coactivators? Provide an example of a coactivator and explain how it regulates gene expression.
→ promote DNA looping by forming bridge bt activator and GTF/RNA pol
ex. mediator- regulates long range interactions bt RTF, GTF, RNA pol
What are effectors? How are they used to regulate activators and repressors?
→ molecules that bind regulatory proteins to change function
effector=change in environment
activator (+) regulation:
gene usually on→ effector arrive→ impair ability of activator to bind DNA→ turn gene expression off
gene usually off→ effector arrive→ enhance ability of activator to bind DNA→ turn expression on
repressor (-) regulation:
gene usually off→ effector arrive→ impair ability of repressor to bind DNA→ turn gene expression on
gene usually on→ effector arrive→ enhance ability of repressor to bind DNA→ turn expression off
How are effectors similar to coactivators? How are they different?
same? both bind TF
different? effector regulate activator/repressor activity while coactivator increase rate tranxn by allowing TF to interact
Bacterial genes use both positive and negative regulation. Describe the two events that need to take place to express bacterial genes at a high level.
(gene usually off)
presence of one effector removed repressor→ no TF is blocking promoter/RNA pol
different effector causes activator to bind DNA→ RNA pol recruited to promoter
What is an operon? What is a polycistronic RNA?
operon→ cluster of prok genes controlled by one promoter
polycistronic RNA→ product of tranxn of an operon, a mRNA that has more than one gene
**one polycistronic RNA can make multiple proteins
Describe the function of the lac operon. Describe the names and functions of the lacZ and lacY genes.
lac operon→ provides genes for bacteria to use lactose as an E source (3 gene w/ 1 promoter)
lacZ: cleaves lactose into glucose and galactose (converts lactose to allolactose)
lacY: imports lactose into cell
lacA
Is the lac operon usually on or usually off in bacterial cells? Provide rationale for your answer.
lac operon usually off: bacteria prefer glucose as E source so the lac operon is only activated when glucose is absent and lactose is present
**controlled by activators and repressors
How does the lacI regulate the lac operon? How does lacO regulate the lac operon?
lacI? lac repressor gene controlled by its own promoter and always expressed
lacO? operator where lac repressor binds when lactose is absent→ looped DNA blocks tranxn
Need to know the meaning of the following genotype designations in bacteria: LacI-, LacIS, LacO-, LacOC, LacP-, Crp-, CrpS
LacI-: LOF mutation of repressor→ cannot bind lacO
LacIS: GOF mutation of repressor→ always bound to lacO
LacO-: LOF mutation of operator→ repressor cannot bind
LacOC: constitutive expression→ lac repressor cannot bind
LacP-: LOF mutation of promoter→ RNA pol cannot bind
Crp-: LOF mutation of cAMP receptor gene→ activator cannot bind
CrpS: GOF mutation of cAMP receptor gene→ activator always bind
Describe the function of CRP and cAMP in regulating the expression of the lac operon.
CRP: activator for lac operon→ effector for this receptor is cAMP
CRP binds cAMP to form a complex→ complex binds CRP site on lac operon→ increase expression
cAMP is an effector molec present when glucose conc low
Describe how effector molecules control positive and negative regulation of the lac operon in the following conditions:
glucose present, lactose absent
glucose present, lactose present
glucose absent, lactose present
Glucose present, lactose absent
cell use glucose for energy
repressor bound to DNA, activator not bound to DNA
no polycistronic mRNA for lac genes is produced
Glucose present, lactose present
cell use glucose and lactose for energy
effector binds repressor and dissociates from DNA
some polycistronic mRNA for lac genes is produced
Glucose absent, lactose present
cell needs lac gene to use lactose as E source
repressor not bound to DNA, different effector binds to activator promoting binding and RNA pol II recruitment
abundant polycistronic mRNA for lac genes is produced
Provide an example for how eukaryotic cells can use a common regulatory protein to regulate several genes.
genes in same pathway controlled by common regulatory proteins→ in positive regulation expression of one activator used to activate multiple genes
**since prok have operons for many gene expression by one promoter, having common proteins conserves energy when diff genes are needed for one pathway
What is combinatorial control? Explain why it is used to regulate a high number of genes in different cell types.
→ genes use common regulatory proteins, but each gene requires a specific combination of reg proteins to activate or inhibit expression
same TF can be used in different complexes to regulate different genes→ multiprotein complexes formed
What is the difference between the major and minor grooves in the double helix? Which is used for transcription factor binding? Why is the groove preferred by transcription factors?
major vs minor groove? differ due to availability for non-covalent interactions
TF interact w/ major groove bc there is more non-covalent interactions
→ more hydrogen bonds available in the major groove compared to the minor groove; providing a much better discrimination between bases
Explain how the shape of transcription factor is important for its function. What is a recognition helix? Why is it common in regulatory transcription factors?
shape? allow specific and stable binding of TF to DNA
recognition helix→ alpha helix in TF that use aa side chains to bind specific DNA seq in major groove
**allows highly specific binding of TF to DNA
What is a helix-loop-helix motif?
protein structure found in many TF→ two alpha helices connected by string of aa (loop)
**only one helix used for recognition and other for dimerization
Why are regulatory sequences often inverted repeats?
inverted repeats signal binding of dimeric TF bc they bind more stably and specifically to DNA
Describe how hormonal/estrogen signaling uses regulatory proteins, regulatory sequences, and effector molecules to regulate gene expression.
activator→ receptor (ER)
effector→ hormone (estrogen)
enhancer→ response element/sequence (ERE)
What is the transcriptional ground state of cells?
→ inherent activity of promoter without regulatory mechanism
What is the transcriptional ground state of prokaryotic cells? Provide a reason for this ground state in prokaryotic cells.
ground state is “on”: RNA polymerase can bind and initiate tranxn in absence of activators and repressors
why? no proteins/chromatin in DNA→ genetic info is open
**regulatory mechanism used to inhibit gene expression
What is the transcriptional ground state of eukaryotic cells? Provide a reason for this ground state in eukaryotic cells and explain why this ground state is advantageous for gene regulation.
ground state is “off”: promoters are inactive in absence of activators
why? due to chromatin structure
advantageous? gene expression more highly regulated
**regulatory mechanism used to activate expression
Compare human and yeast promoters. What is similar? What is different? What is a UAS?
human→ many enhancers (binding sites for activators) located upstream, downstream, close, or near TSS
yeast→ few upstream activator sequences (UAS) as binding sites for activators
same? multiple regulatory sequences for each gene
different? only UAS for yeast while humans have both
Explain the activation and repression for most eukaryotic genes. How are these mechanisms related to the ground state in eukaryotic cells?
activation? coactivators provide bridge bt activators and RNA polymerase
repression? corepressors prevent activators from interacting with RNA polymerase
Describe gene activation and repression in yeast using the following terms: corepressor, general transcription factors (GTFs), mediator, activator, RNA pol II initiation complex.
activation? GTF bind promoter → activators bind UAS→ mediator (coactivators) bridges activator and RNA pol II into initiation complex
repression? corepressor binds activator and disrupt interactions with initiation complex
What are GAL genes used for in yeast? Why are they regulated?
GAL gene used to metabolize galactose as an E source
regulation? gene is activated in presence of galactose bc its prioritizes glucose as E source
**regulated by common proteins even with genes on different chromosomes
Describe how the GAL genes are repressed and activated due to changing environmental conditions. In your description, include the terms Gal4p, Gal80p, Gal3p, and galactose.
Gal4p: activator, bind UAS site
Gal80p: corepressor, binds activator
Gal3p: bind Gal80p and galactose (when galactose and ATP are present)
repression? low galactose concentration, Gal80p binds Gal4p and prevents it from interacting with initiation complex
activation? high galactose concentration and Gal3p binding to Gal80p causes conformational change→ allow Gal4p interacts with initiation complex
Why are many regulatory transcription factors homodimers? Why is it advantageous for eukaryotic cells to use transcription factors that are heterodimers?
homodimer→ one protein with two identical subunits: provide more protein-DNA interactions at regulatory sequences
heterodimers→ one protein with two different subunits: allow euk cells to create many functional tranxn factors from small number of individual proteins
What is a reporter gene? What type of information is provided when reporter genes are used in an experiment?
→ gene attached to gene of interest or regulatory sequence to measure gene expression
used to study location of protein within cell or entire organism when fused to gene of interest
used to analyze function of regulatory sequence (promoters and enhancers)
What are some examples of reporter genes?
GFP
LacZ
Famous experiment where scientists were investigating whether different domains of regulatory proteins could be separated but still functional:
Describe the fusion protein in this experiment.
Describe the reporter gene in this experiment.
Describe the results and overall conclusion of the experiment
fusion protein consisted of a LexA DNA-binding domain (prok promoter) and Gal4p activation domain (euk activator binding site)
two reporter genes used, one with LexA binding site and one without LexA binding site→ measure expression of LacZ (see if Gal4p can interact w/ GAL1 promoter)
tranxn activation and DNA-binding domain can be separated and function independently of one another
result: DNA-binding domain and the activation domain can function independently yet cooperatively within a fusion protein
What is epigenetics?
→ study if heritable changes in gene expression that occur without alterations to the DNA sequence
What are epigenetic modifications? How are they similar to DNA mutations? How are they different?
epigenetic mods? chemical mods to DNA and histones that regulate tranxn
same? heritable + alter gene expression + cause disease
different? epigenetic mod do not alter DNA sequence while DNA mutations do. DNA mutations can be repaired but mods cannot be corrected.
Why is the agouti mouse a good example of how epigenetics can be used to regulate a trait of an organism?
agouti mouse: coat color controlled by agouti gene→ gene regulated by methylation
orange and brown agouti mice can be genetically identical→ same DNA seq at gene but different epigenetic mod= diff characteristics
Compare DNA methylation and histone modifications. How are they similar? How are they different?
same? influences gene expression
DNA methylation: chemical mod to DNA
Histone mod: chemical mod to histone **work with chromatin remodeling complexes
Need to be familiar with the following terms/concepts: histones, nucleosomes, N-terminal tail, acetylation at H3K4, H3K9, and H3K27, methylation at H3K4 and H3K9.
histones: proteins that DNA wraps around for compaction in euk cells
globular domain→ DNA wraps around
N-terminal tail→ protrusions from ^ that allow intermolecular contact for compaction regulation and can be chemically modified
nucleosomes: octamer of histones
acetylation: associated w/ euchromatin aka tranxn activation
H3K4, H3K9, and H3K27→ euchromatin when acetylated
HAT→ adds acetyl gp lysine to aa @ N-terminal
HDAC→ remove acetyl gp lysine to aa @ N-terminal
methylation:
H3K4me= euchromatin/activation
H3K9me & H3K27me= heterochromatin
methyltransferase→ adds methylation to lysine
demethylase→ removes methylation from lysine
What is a bromodomain? What is a chromodomain?
bromodomain: protein domain that recognize and binds to acetylated lysine residues in histones and other proteins
chromodomain: protein domain that recognize and binds to methylated lysine residues in DNA
Describe the relationship between histone modifications and chromatin remodeling complexes.
histones subunits with acetylation after replication recruit bromodomain-containing CRC→ HAT subunits in CRC propagate acetylation to neighboring histones to promote euchromatin OR activation of gene by nucleosome rearrangement by CRC
histone mods recruit CRC through bromodomain recognition
CRC uses HAT activity to spread acetylation
Are histone modifications heritable? In other words, can they be maintained after DNA replication? If so, provide examples.
yes!
some histone core subunits remain bound to DNA during replication
CRC used to recognize and maintain the modifications using bromodomain and HAT
What is DNA methylation? Where does DNA methylation occur in eukaryotic cells?
→ adding a methyl group to a base (cytsoine) to form 5-methyl cytosine
location? CpG symmetric dinucelotides→ same sequence in same orientation
**CpG islands: cluster of CpG dinucleotides
What is the function of a DNA methyltransferase (DNMT)?
enzyme used to methylate DNA
ex. cytosine→ 5-methylcytosine
What effect does DNA methylation at promoters typically have on the expression of a gene?
methylation in or around promoter typically inhibits gene expression
Describe two ways DNA methylation can regulate expression of genes in eukaryotic cells.
prevents binding of GTF to a promoter
no GTF=no RNA pol binding= no tranxn initiation
prevents binding of RTF to regulatory sequence
no activator bound to enhancer= tranxn decrease
What is maintenance methylation?
DNMT localizes to replication fork during DNA replication and methylates CpG dinucleotides on daughter strand of DNA if CpG site on template strand is methylated
CpGs are symmetric
replication is semiconservative
Describe why DNA methylation is important for the development of a multicellular organism.
bc of cell differentiation!
→ diff cell types have diff DNA methylation patterns (established during early development) this allows them to have unique expressions and specified roles
**methylation patterns are maintained during cell division→ stable and heritable
Explain what is meant by the phrase “a human body has one genome but many epigenomes”?
one genome bc different cell types in humans have the same DNA sequence
many epigenomes bc different cell types have different DNA methylation patterns
Experiment 1 analyzed young and old twins→ describe the experimental design and the results
study looked at differences in DNA methylation bt identical twins
young twins had similar DNA methylation patterns while old twins had different DNA methylation patterns
conclusion? epigenetic mods are altered by environment throughout the life of an individual
Experiment 2 used the agouti mouse→ describe the experimental design and the results
can maternal diet influence epigenetic mods in developing fetus?
pregnant female agoui mice with orange fur (unmethylated) were fed low folate diet or rich folate diet (DNA methyl donor) during pregnancy
since coat color is controlled by DNA methylation→ orange females gave birth to brown offspring through high folate diet
conclusion? diet of pregnant mom can influence epigenetic mods of developing fetus
demonstrated that the environment can modify epigenetic modifications in an individual
What is the function of ribosomes within cells? What are the functions of the small and large subunits of ribosomes?
ribosome: large protein complex that synthesizes proteins using genetic information within mRNA
small subunit: matches tRNAs to codons on mRNA
large subunit: generates peptide bonds between amino acids
What are ribosomes made of? Proteins, RNA, or both? If they have RNA, is it in the small subunit, large subunit, or both subunits?
ribosome complex is made of many proteins and at least one ribosomal RNA (rRNA) in each subunit→ for both euk and prok
prok:
large (2): 23S and 5S
small (1): 16S
40:60 (protein:RNA)
Ribosome biogenesis is a multi-step process. Identify some critical steps that must occur during this process in prokaryotic cells.
transcribe mRNA for ribosomal proteins
translate the mRNA to make ribosomal proteins
transcribe and process rRNA primary transcript
assemble functional ribosome using mature rRNA and ribosomal proteins
**assembly occurs while rRNA is being transcribed by RNA polymerase
Describe the use of operons to produce prokaryotic ribosomes.
50 ribosomal proteins are regulated by ~15 operons→ some operons have genes for small ribosomal proteins, some for large ribosomal proteins, and some operons have genes for both
In class, we discussed how a specific protein, S8, regulates expression of ribosomal proteins. Describe how prokaryotic cells use this protein to shut down gene expression when the cell has made enough ribosomes. Can it bind to mRNA, rRNA, or both?
proteins S8 is produced by translation→ can bind both mRNA and rRNA but has greater affinity for rRNA
when concentration S8>rRNA, S8 its own binds polycistronic mRNA
binding of S8 to mRNA blocks translation and induces mRNA degradation with ribonucleases
negative feedback loop→ S8 controls the translation of the mRNA that codes for more ribosomal proteins including itself (S8)
Describe the process of ribosome biogenesis for the small and large subunits in prokaryotes. Start at transcription of rRNA then explain how ribosomes are made with mature rRNAs and ribosomal proteins.
ribosome assembly occurs while rRNA is being transcribed by RNA polymerase
after assembly, rRNA is process
pre-rRNA transcript has bases modified by enzymes— mods allow folding
pre-rRNA transcript is cut by ribonuclease to liberate 16S, 23S, 5S rRNA
mature 30S and 50S subunits combine to make functional 70S ribosomes
Where are ribosomes located in a eukaryotic cell? Are all ribosomes the same in a eukaryotic cell?
80s ribosomes are observed in cytoplasm and rough ER
mitochondrial ribosomes (mitoribosomes) are similar to prok 70S
Does the eukaryotic ribosome have ribosomal RNA? If so, where is the RNA located in the ribosome? Small subunit, large subunit or both subunits?
both large and small subunits of euk ribosomes have rRNA
large (3): 28S, 5.8S, 5S
small (1): 18S
50:50 (protein:RNA)
How is ribosome biogenesis in eukaryotes similar to prokaryotes? How is it different?
same?
transcribe rRNA and mRNA for ribosomal proteins
process rRNA primary transcript
assemble functional ribosome using mature rRNA and ribosomal proteins
different?
euk→ some rRNA transcribed in nucleolus and some in nucleus, by different RNA polymerases
euk→ snoRNAs used to cut pre-rRNA and modify bases in nucleolus
euk→ ribosomal proteins bind processed rRNA in nucleolus and exported to cytoplasm for translation
Explain how all three RNA polymerases are important for ribosome biogenesis in eukaryotes.
RNA pol II: makes all mRNA→ all ribosomal proteins
RNA pol I: makes most rRNA→ 45S pre-rRNA turns into 18S, 5.8S, and 28S rRNA
RNA pol III: makes some rRNA and all tRNA→ 5S rRNA made
What is the nucleolus? Describe its location and function.
→ area within nucleus that produces ribosomes
Describe the 45S (aka 47S) ribosomal gene in eukaryotic cells? Where is this gene located? Describe its location in the genome and within the cell. What is this gene used to produce?
45S rRNA is produced in the nucleolus by RNA pol I
euk cells have over 100 copies of 45S rRNA in genomes (repeated over several chromosomes)
sections of chromosome containing gene are recruited to nucleolus
45S→ processing to make 18S, 5.8S, and 28S rRNA
What are snoRNPs and snoRNAs? Why are they important for ribosome biogenesis?
→ enzyme contains protein and RNA used to modify bases in 45S pre-RNA after tranxn
mods are extensive and occur at specific bases
mods increase non-covalent interactions of bases→ help with folding of rRNA into mature ribosomes
once modified→ 45S pre-RNA is cleaved to produce 3 rRNA’s (18S, 5.8S, 28S)
Describe the process of ribosome biogenesis for the small and large subunits in eukaryotes. Start at transcription of rRNA then explain how ribosomes are made with mature rRNAs, ribosomal proteins, snoRNPs, and assembly factors. Make sure you explain how all three polymerases contribute to this process.
45S pre-RNA is transcribed in nucleolus and processed/modified by snoRNP to mature, 5S rRNA is made in nucleus, ribosomal proteins made by translation of mRNA in cytoplasm
5S rRNA and proteins are imported into nucleolus
small and large ribosomal proteins bind to rRNAs
90S ribosome forms in nucleolus→ contains small and large subunits, rRNAs, snoRNPs, and assembly factors
inactive complex ^ imported to cytoplasm where 40S and 60S become functional subunits and can perform protein synthesis→ release assembly factors
**ribosomal biogenesis ends in the cytoplasm
What is the difference between the transcriptional start site and the start codon? Describe their locations.
TSS: first base that is transcribed @ DNA
start codon: first amino acid translated @ mRNA
What are untranslated regions? Describe their locations and provide at least one function of an untranslated region.
→ sequence in mRNA that does not contribute to protein— space between TSS and start codon
function? recognized regions of mRNA by ribonucleases during mRNA degradation
What are codons? Need to be able to identify start and stop codons (all three) in an RNA sequence.
codons: sequence of three nucleotides in DNA or RNA that correspond to a single amino acid during protein synthesis
**three letter= one amino acid
start→ AUG
stop→ UGA, UAA, UAG
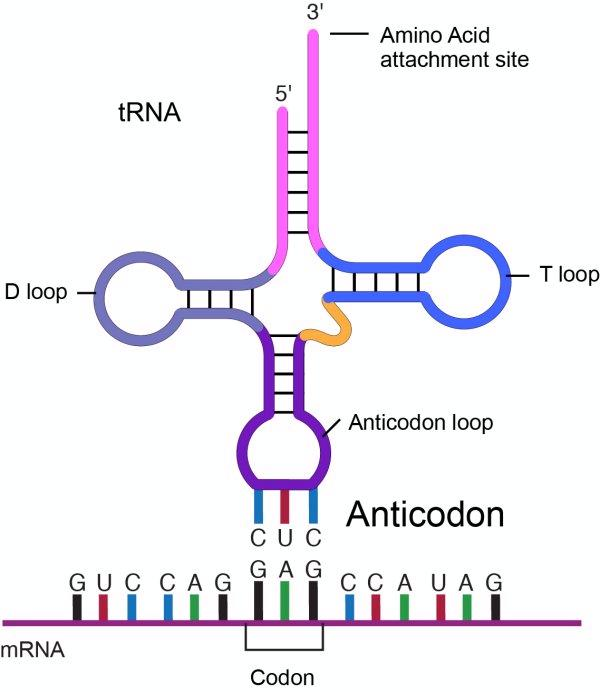
What are transfer RNAs (tRNAs)? What are anticodons?
tRNA: translate genetic sequence in mRNA to amino acid sequence (protein)
anticodon: three base sequence in tRNA that is complementary to a codon in mRNA
What is the genetic code? Why is the genetic code “degenerate”?
→ set of rules where information encoded within genetic material (DNA or mRNA) is translated into protein by living cells (universal)
degenerate? one amino acid often uses several different codons (64 codonds w/ only 20 amino acids)
What are wobble bases? Describe their location. How do cells use the wobble base to make the genetic code degenerate?
wobble bases→ base in codon and anticodon that do not always follow base-pair rules (more flexible bp)
examples?
tRNA w/ G or U in 5’ position can recognize two codons
tRNA with I (inosine) in wobble position can recognize three codons
contribute to degenerate? allow multiple base pairing to code for one amino acid
Describe (or draw) an anticodon binding to a codon using 5’ and 3’ ends.
Why do some amino acids require multiple tRNAs to recognize all codons?
amino acids with more then 3 different codons need more then one tRNA bc this maxes out the wobble base flexibility (with inosine)
ex. codon families like Val or Ser (6 codons)
Describe at least one advantage for having a degenerate genetic code. We discussed two in class.
cells require fewer tRNAs→ greater efficiency and use of resources
silent mutation increase→ cells are able to absorb more mutations
What is the reading frame of a gene? Why is choosing the correct reading frame important? How do ribosomes choose the right reading frame?
reading frame: sequence of codons used for proteins synthesis
importance? only the correct reading frame with produce the sequence to synthesize the correct protein
ribosome contribute? start codon on mRNA indicates correct reading frame for ribosome
What is the initiation complex in translation? Describe the sequence of events that leads to the formation of the initiation complex.
initiation complex: ribosome + mRNA + initiator tRNA form a complex around start codon to initiate translation
What is an open reading frame?
→ long segment of DNA nucleotides in a gene sequence with no stop codons
**statistical anomaly→ if mRNA is translated with the wrong reading frame, a stop codon is statically likely to arise
Describe the processing and the structure of tRNAs. Why are the bases modified?
DNA transcribed and RNA transported to cytosol
tRNA processing
5’ and 3’ ends cut by RNase
bases modified→ helps with folding and wobble flexibility
CCA added to 3’ end → aa binding site
introns spliced (if needed)
3D structure= letter-L
What are two important functions of tRNAs?
recognize correct codon sequence in RNA (@ anticodon)
carry aa specific for the codon (@ 3’ end)
**very important that tRNA connect to right aa
What is the function of aminoacyl tRNA synthetases? Describe how these proteins perform their function in two steps.
ARS→ protein that attaches amino acids to 3’ end of mature tRNA in cytosol
how?
ATP and aa bind to ARS, ATP is used to link aa to AMP forming aminoacyl-AMP
tRNA binds ARS, aa is transferred from AMP to tRNA to form aminoacyl-tRNA→ tRNA is now charged
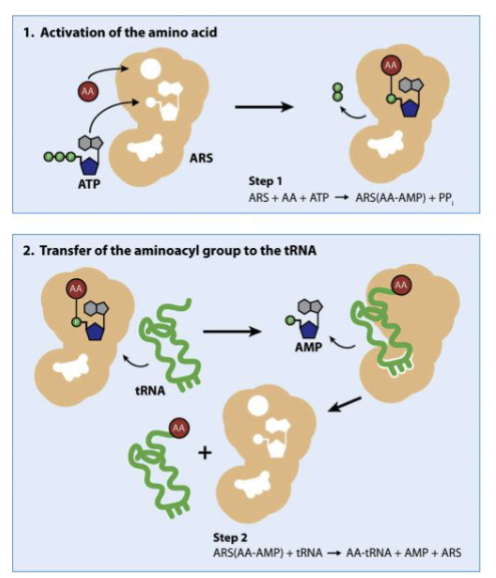
What is an aminoacyl tRNA?
→ tRNA with an amino acid attached at the 3’ end
“charged tRNA”
Identify the three sites in a ribosome that are used for translation and describe the function of each site.
Aminoacyl site: binding site for charged aminoacyl tRNA
Peptidyl site: site where polypeptide chain is attached to peptidyl tRNA
Exit site: exit site for uncharged tRNA
Identify and describe the three stages of translation used by prokaryotes and eukaryotes.
initiation:
ribosomal subunits, mRNA, and initiator tRNA form initiator complex @ start codon
reading frame established
initiator tRNA is in P site
codon for next amino acid enters A site
elongation:
ribosome moves down mRNA and adds amino acids to growing chain by forming peptide bonds
charged tRNA enter A site and add amino acid to chain
termination:
ribosome reaches stop codon
release factors recruited to A site→ release polypeptide
mRNA-ribosome complex fall apart→ recycle
What is a Shine-Dalgarno sequence? What is its function? Where is it located?
shine-dalgarno sequence→ consensus sequence on 5’ end of mRNA that binds to 16S rRNA in small subunit of ribosome
function? this is how the small subunit binds mRNA in prok cells
What is the initiator tRNA? What is its function?
initiator tRNA: binds start codon at initiation
Methionine only has one codon. Why do all organisms have two tRNAs for methionine?
initiator tRNA: binds to start codon at at P site
elongator tRNA: codes for methionine in A site
Describe the aminoacyl tRNA for the start codon. How is it different from the aminoacyl tRNA that codes for internal methionine residues (the Met residues not in start codon)?
initiator created by adding a formyl group to the methionine already attached to a tRNA
What is a GTP-binding protein? How is the activity of a GTP-binding protein regulated?
GTP-binding protein: has domain for binding to GTP (active) or GDP (inactive)
regulation?
GEF→ promotes binding of GTP
GAP→ promotes hydrolysis of GTP
Describe how three different initiation factors promote initiation of translation in prokaryotic cells. Why is GTP hydrolysis important for the initiation stage?
IF-1 binds small subunit and blocks tRNA binding
IF-3 binds small subunit and prevents premature activation of ribosome
**rRNA binds Shine-Dalgarno
IF-2 (GTP-binding protein) w/ GTP attached recruits initiator tRNA to start codon in P site of small subunit
**GTP hydrolyzed by ribosome→ IF released, large subunit binds, initiation complex formed
Describe the three steps of elongation. Why is GTP hydrolysis important for the elongation stage?
aminoacyl-tRNA binds to A site in ribosome using base-pairing between anticodon and codon
aminoacyl-tRNA binds EF-Tu w/ GTP attached→ delivers tRNA to A site and only hydrolyzes if correct bp match
EF-T is a GEF for EF-Tu, attaches GTP so it can bind another tRNA
peptide bond formed bt amino acid in P and A sites→ growing polypeptide transferred to A site
peptidyl transferase rxn @ large subunit
translocation shifts tRNA without amino acid to E site, tRNA w/ polypeptide to P site, and new codon in A site
EF-G-GTP (mimics EF-Tu-aminoacyl tRNA) binds A site and hydrolyzes to EF-G-GDP which promotes translocation
cycle goes on and on!
What is accommodation in protein translation? How is it important in ensuring the accuracy of translation?
accommodation: rotation of tRNA into position for peptide formation through correct codon-anticodon base pairing
correct→ ribosome converts EF-Tu-GTP to EF-Tu-GDP
incorrect→ EF-Tu-GTP released from A site
**proofreading mechanism
What are the two goals of termination? How do prokaryotic cells accomplish each goal? Why is GTP hydrolysis important for the termination stage?
release of polypeptide
release factors (RF) work to promotes release of polypeptide chain→ 1st RF release polypeptide and 2nd RF uses GTP hydrolysis to removed 1st RF from A site
dissociation (and recycling) of subunits, mRNA, and tRNA
ribosome recycling factors (RRF) has same shape as tRNA and fit A site with stop codon→ recruit EF-G-GTP to A site
translocation through hydrolysis→ RRF @ P site and IF-3 binds small subunit to break ribosome
Describe how the beginning of initiation in eukaryotes is similar to prokaryotic cells. How is it different?
same?
initiation factors separated and prevent tRNA binding to A site
establish reading frame with start codon
different?
euk 43S pre-initiation complex forms prior to mRNA bindings
Shine-Dalgarno sequence vs recognition of the 5' cap
IF vs eIF
initiator tRNA w/ formyl gp vs without
Compare and contrast initiator aminoacyl tRNAs used by eukaryotic and prokaryotic cells. How are they similar? How are they different?
same? two tRNA for met, initiator and elongator
different? euk no formyl gp for initiator tNA
How is the mRNA recruited to the ribosome in eukaryotic cells? Is this the same mechanism used in prokaryotic cells?
prok→ shine-dalgarno sequence used to recruits mRNA to ribosome
euk→ 5’ cap used to recruit mRNA to ribosome (by eIF4F)
eIF4F is an important initiation factor for eukaryotic translation. Describe the different functions of eIF4F.
complex of proteins that binds 5’ cap and uses ATP to recruit mRNA to preinitiation complex to for 48S
functions?
bind 5’ cap
connect complex to small subunit
hydrolyze ATP to complete attachment (~glue)
What happens after eIF4F joins the pre-initiation complex to create the 48S particle?
eIF4F scans 5’ to 3’ direction to find start codon after 5’ cap→ establish reading frame
What process allows small and large subunits to come together to form a functional ribosome in eukaryotic cells? Is this process similar or different from prokaryotic cells?
small subunit and initiator tRNA bound at start codon
large subunit joins after IF dissociate through hydrolysis
large ribosomal subunit binds only after the mRNA and initiator tRNA have attached to the small ribosomal subunit and IF’s have left
Eukaryotes: small subunit first scans the mRNA to locate the start codon, and only then does the large subunit join.
Prokaryotes: small subunit aligns with the mRNA’s Shine-Dalgarno sequence, positioning the start codon directly, and the large subunit joins without any scanning