PCR and Southern blotting
1/13
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
14 Terms
What changes can you do when PCR has no target bands
For little to no target band:
1. Check your math
2. Verify all components are present
3. Verify primer design
4. Consider if DNA sample has inhibitors (amplified successfully in other reactions?)
5. Lower annealing temperatures
6. Add more template or cycles
7. Add more magnesium
8. Use PCR enhancers or higher denaturation temps for GC-rich targets
What can you do if your PCR has non-specific products
For non-specific products:
1. Increase annealing temperature
2. Reduce primer concentrations (especially if there are abundant primer dimer complexes)
3. Shorten annealing and extension times
4. Reduce cycles or template
5. Reduce magnesium
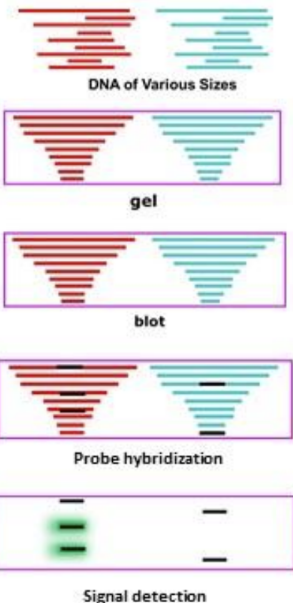
What are the steps to southern blotting
Southern blot steps:
Restriction enzyme digest
DNA is isolated and then cut
Electrophoresis of agarose gel
Fragments are separated by gel electrophoresis, denatured in the gel
Transfer DNA to nitrocellulose membrane (blotting)
Fragments are transferred to a solid support membrane
Hybridize probe to plot
DNA fragments on the membrane are exposed to a labeled probe that is complementary to the region of interest
Probes are usually larger (allow to see individual bands)
Wash blot
Detection of probe signal
The signal of the probe is detected to indicate the presence or absence of the sequence of interest
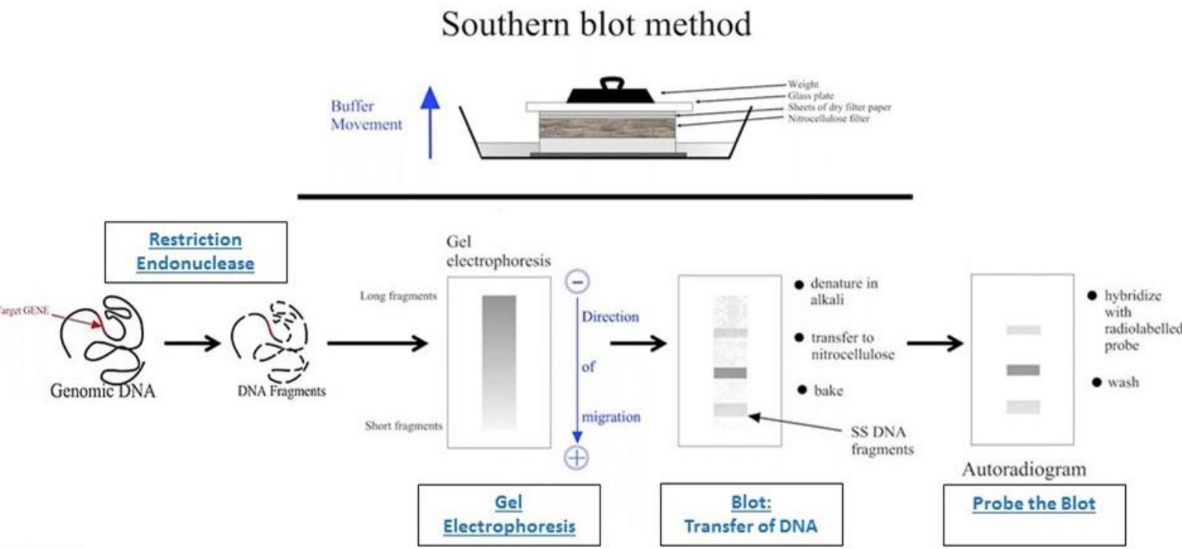
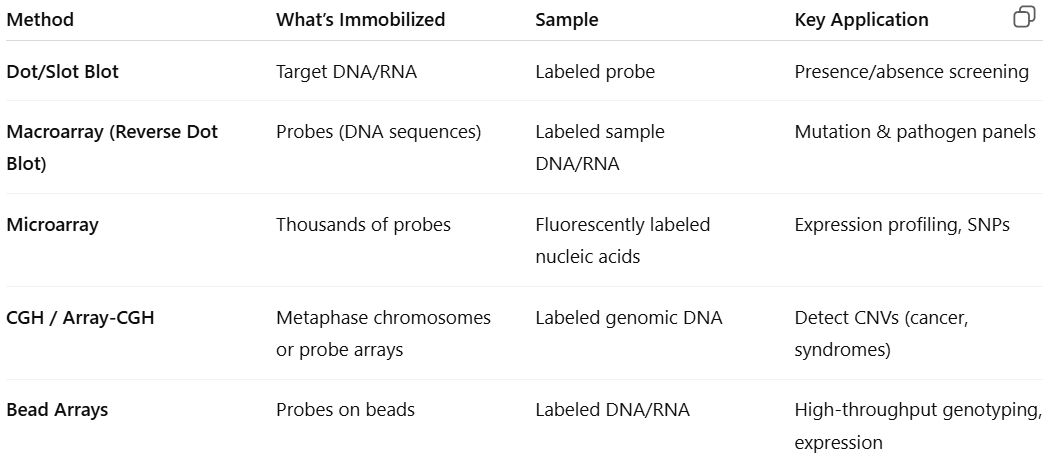
What is dot/slot blots
Dot/slot blots: target DNA/RNA is deposited directly on the membrane by means of various devices (ex: vacuum system)
Applied to expression, mutation, and amplification/deletion analyses
Determination of size is not required
Most efficient on less complex samples
Dot blots = target is deposited in a circle or dot
More useful for multiple qualitative analyses where many targets are being compared (mutational screening)
Ability to test and analyze larger numbers of samples at the same time
Slot blots = target is deposited in an oblong bar
More accurate for quantification by densitometry scanning because they eliminate the error that may arise from scanning through a circular target
What is macroarrays (reverse dot blot)
Macroarrays (reverse dot blot): many different unlabeled probes are immobilized on the membrane, and the test sample is labeled for hybridization with the immobilized probes
a known sequence is immobilized at a known location on the blot and the amount of sample that hybridizes to it is determined by the signal from the labeled sample
Limited by the area of the membrane and the specimen requirements
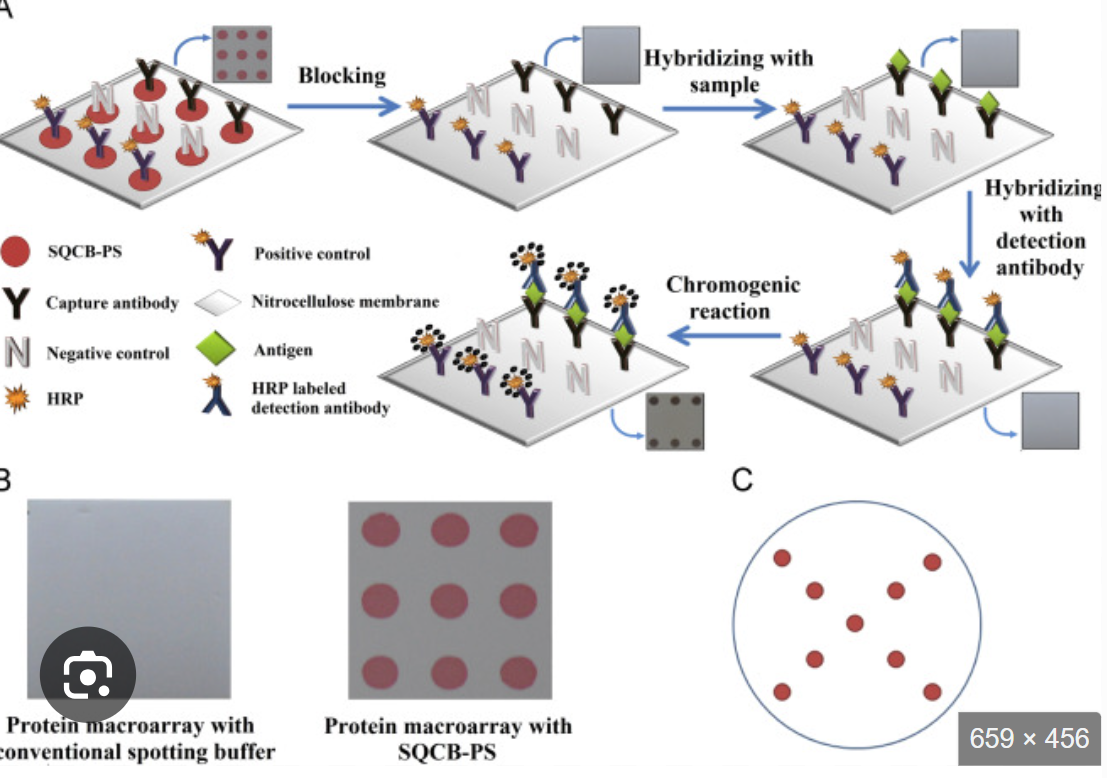
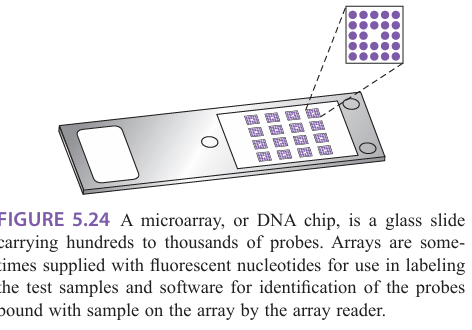
What is microarrays
Microarrays:
Tens of thousands of targets could be screened simultaneously in a very small area by miniaturizing the deposition of droplets
Array targets immobilized on glass slide
Targets can be DNA, RNA, or protein
Requires fluorescent reader and analysis software
Probes are immobilized on a solid support
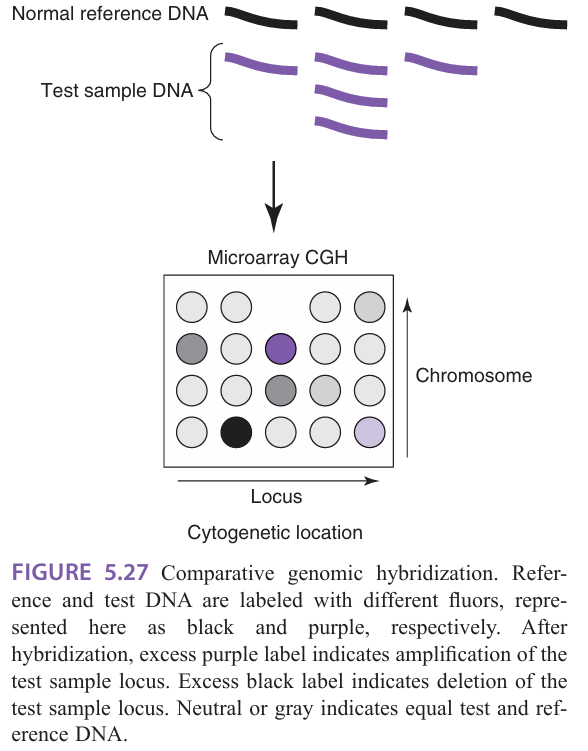
What is comparative genome hybridization (CGH)
Comparative genome hybridization (CGH): designed to test DNA
Used to screen the genome or specific genomic loci for deletions and amplifications
Genomic DNA is isolated, fragmented, and labeled for hybridization on the chip
Provides higher resolution and more defined genetic information than traditional cytogenetic analysis
Limited to the analysis of loci represented on the array
Advantage = can be performed on fixed tissue and limiting samples
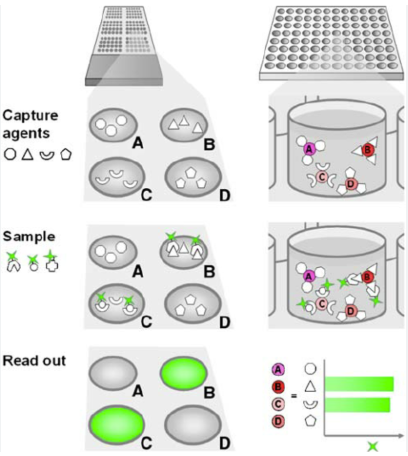
What is bead arrays
Bead arrays: immobilize probes with beads, allowing hybridization of the targets in the bead suspension
Used for protein and nucleic acid targets
Available for infectious disease and tissue typing
Beads are color coded with a particular shade of red fluorescent dye so you can distinguish specific probes carried on different beads
What are the concepts and applications of the following hybridization method: Dot/slot, macroarrays, microarrays, CGH, and bead arrays
look at pic
What are applications of southern blots
Applications of southern blots: (southern blotting is “old school”)
Genetics, oncology (translocations, gene rearrangements)
Detection of repeat expansions (FXS, Huntington)
Typing/classification of organisms
Cloning/verification of cloned DNA
Forensic, parentage testing (RFLP, VNTR)
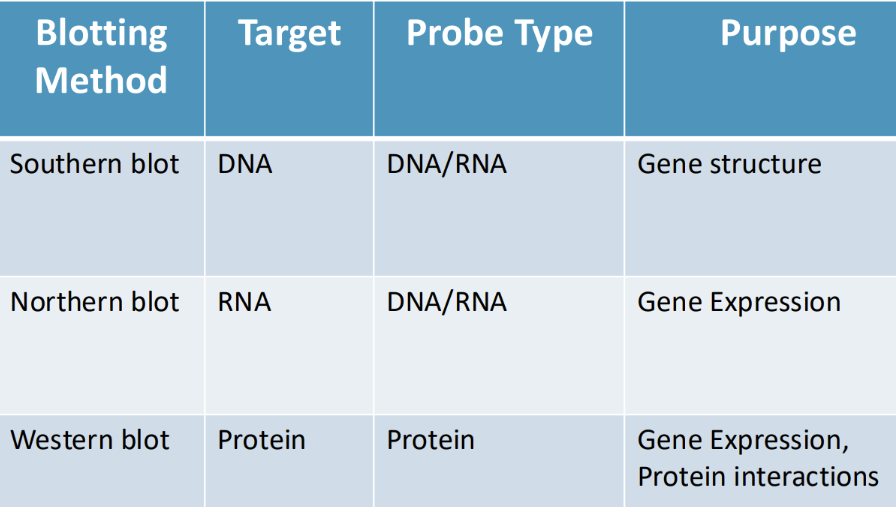
What is the purpose and probe type of Southern, Northern, and Western blots
picturezzzzzz
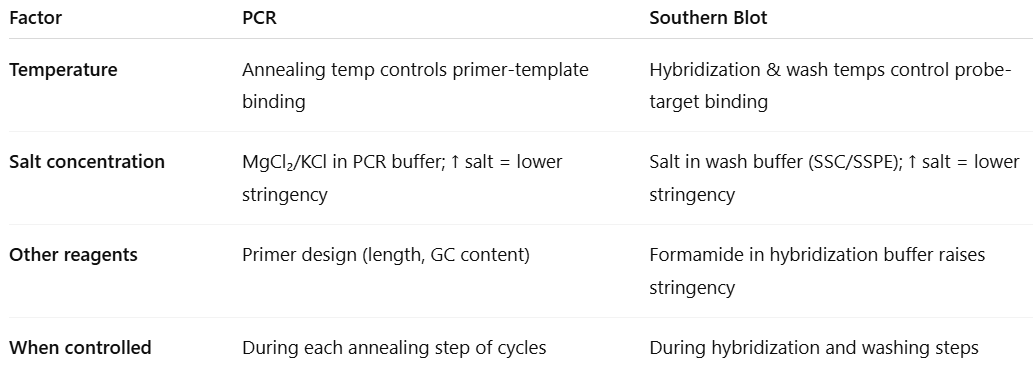
What are factors that affect stringency in blotting
Factors that affect stringency:
Temperature of hybridization
High temperatures = higher stringency
Low temperatures = lower stringency
Wash temperature
high temperatures = higher stringency
low temperatures = lower stringency
Hybridization time
more time = lower stringency
less time = higher stringency
Wash time
lower time = lower stringency
higher time = higher stringency
Salt concentration of hybridization buffer
Keep hybridization solution low
High salt = lower stringency
Lower salt = higher stringency
Concentration of denaturant (formamide) in the buffer
Formamide lowers the optimal hybridization temperature
More formamide = more stringency
Length and nature of probe
Long probe or high GC bases = binds in more stringent conditions
Require longer hybridization times
Short probe or high AT bases = binds in lower stringent conditions
Require lower hybridization times
Increased probe concentration = increased sensitivity of analysis
Ideal conditions = calculated with Tm of probe sequence (and Cot)
How does stringency relate to probe binding
High stringency = more demanding of probe/target complementarity and length
If too high, the probe will not bind to target
Low stringency = more forgiving binding
If too low, the probe will bind to unrelated targets
How is stringency in PCR and southern blotting similar
look at the pic