DNA Damage and Repair Mechanisms
1/50
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
51 Terms
Oxidative stress
A condition characterized by excessive free radicals, which can lead to cellular damage.
8-Oxo-2'-deoxyguanosine (8-oxo-dG)
A common form of oxidative DNA damage that results from the attack of hydroxyl radicals on guanine.
Thymine Dimer
A type of DNA damage induced by UV light where two thymine bases bond covalently.
Non-Homologous End Joining (NHEJ)
An error-prone DNA repair mechanism that directly joins broken DNA ends.
Homology-Directed Repair (HDR)
A precise DNA repair mechanism that uses a homologous sequence as a template.
Mismatch Repair
A DNA repair process that corrects base-pairing mismatches during DNA replication.
DNA polymerase
An enzyme that synthesizes new DNA strands by adding nucleotides to a template strand.
DNA Ligase
An enzyme that seals nicks in the DNA backbone during repair processes.
Photorepair
A direct repair process that reverses thymine dimers caused by UV light using DNA photolyase.
Methylguanine Methyltransferase (MGMT)
A 'suicide enzyme' that repairs O6-methylguanine lesions by transferring the methyl group to itself.
DNA Damage
Genetic alterations caused by environmental, chemical, and biological agents that affect the integrity of the genome.
Mutation
A change in the DNA sequence of a genome, which can lead to variations in protein encoding.
Point Mutation
A mutation that involves a change in a single nucleotide, which can be a transition or transversion.
Transition Mutation
A point mutation that replaces a purine with another purine or a pyrimidine with another pyrimidine.
Transversion Mutation
A point mutation that replaces a purine with a pyrimidine or vice versa.
Nonsense Mutation
A mutation that creates a premature stop codon, resulting in incomplete and often nonfunctional protein.
Silent Mutation
A mutation that does not change the amino acid sequence of the resulting protein.
Insertion Mutation
A mutation where extra nucleotides are added to the DNA sequence, which can cause a frameshift if not in multiples of three.
Deletion Mutation
A mutation where nucleotides are removed from the DNA sequence, potentially causing a frameshift if not in multiples of three.
Oxidative Stress
A condition characterized by elevated levels of reactive oxygen species, which can cause DNA damage.
Depurination
The loss of a purine base from the DNA, leading to potential mutations and strand breakage.
Alkylation
The addition of alkyl groups to DNA bases, potentially altering base-pairing and leading to mutations.
UV-Induced Thymine Dimer
A type of DNA damage where adjacent thymine bases bond covalently, disrupting DNA structure and replication.
Intercalating Agents
Substances that insert themselves between DNA bases, distorting the DNA structure and potentially causing frameshift mutations.
DNA Repair Mechanisms
Biological processes that correct DNA damage to maintain genomic integrity.
Huntington’s Disease
A genetic disorder caused by the expansion of CAG repeats in the HTT gene, leading to neurodegeneration.
Biological Causes of DNA Mutations
Inherent processes such as replication errors and slippage that can introduce mutations into the genome.
Chemical Agents of DNA Damage
Substances that can alter DNA structure and function, leading to mutations.
Environmental Agents of DNA Damage
External factors such as radiation, tobacco, and dietary components that cause DNA damage.
Chemical Mutagens
Substances that induce mutations in DNA, such as alkylating agents, intercalating agents, and base analogs.
What are the 3 major types of mutations?
1. Base Substitution: A change in a single nucleotide, replacing one base with another.
2. Insertion: The addition of one or more nucleotides into a DNA sequence.
3. Deletion: The removal of one or more nucleotides from a DNA sequence.
What are transitions vs transversions, and how can you recognize them in a DNA sequence?
Both are types of point mutations:
Transition Mutation: Involves replacing a purine with another purine (e.g., A \leftrightarrow G) or a pyrimidine with another pyrimidine (e.g., C \leftrightarrow T). These maintain the same base chemical class.
Transversion Mutation: Involves replacing a purine with a pyrimidine or a pyrimidine with a purine (e.g., A \leftrightarrow C, T \leftrightarrow G). These switch between base chemical classes.
Recognition: To identify them, observe whether the substituted base belongs to the same chemical class (transition) or a different chemical class (transversion) compared to the original base.
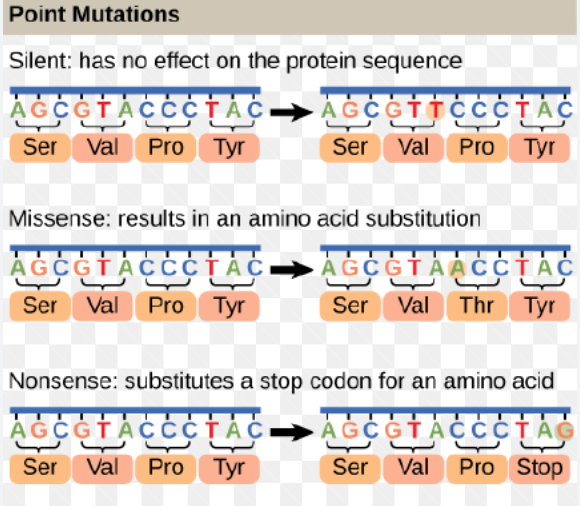
What are the potential consequences of coding sequence base substitutions (silent, nonsense, missense)?
Base substitutions in a coding sequence can lead to different outcomes for the resulting protein:
Silent Mutation: A base change that does not alter the amino acid sequence, often due to the degeneracy of the genetic code (e.g., replacing a codon with another that codes for the same amino acid). These typically have no effect on protein function.
Missense Mutation: A base change that results in a codon specifying a different amino acid. The impact on protein function can vary from negligible to severe, depending on the biochemical properties of the new amino acid and its location in the protein.
Nonsense Mutation: A base change that converts an amino acid-specifying codon into a premature stop codon. This leads to the production of a truncated, often non-functional protein.
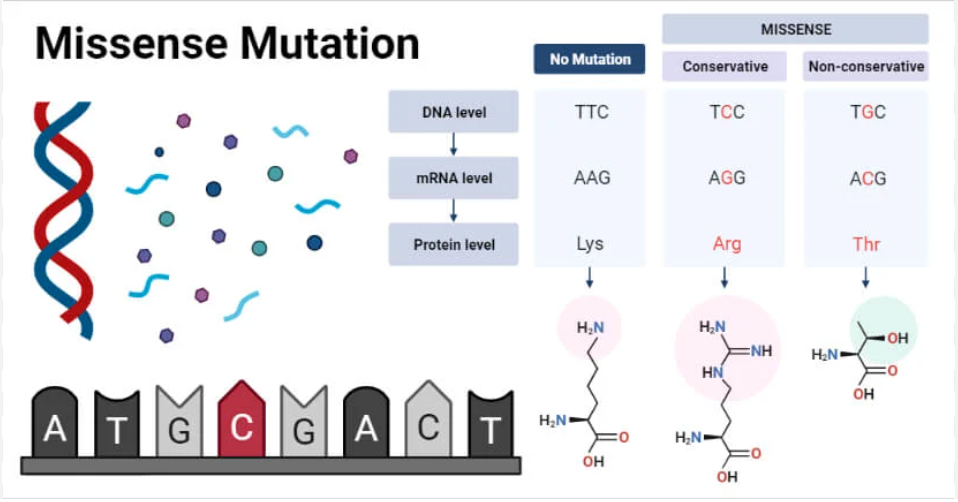
What is the difference between a conservative missense mutation and a non-conservative missense mutation?
Both are types of missense mutations, categorized by the nature of the amino acid substitution:
Conservative Missense Mutation: The substituted amino acid has biochemical properties (e.g., size, charge, polarity) similar to the original amino acid. This often results in a protein that retains some or most of its original function.
Non-conservative Missense Mutation: The substituted amino acid has significantly different biochemical properties compared to the original amino acid. This type of mutation is more likely to cause a major change in protein structure and function, potentially leading to a non-functional protein or disease.
Which codon position is most likely to cause a silent mutation?
The third codon position is most likely to cause a silent mutation because of the degeneracy of the genetic code, meaning multiple codons can specify the same amino acid. While changes here often don't alter the amino acid, missense or nonsense mutations can still occur depending on the specific nucleotide change.
What is a frameshift mutation, and what are its consequences?
A frameshift mutation is a type of gene mutation caused by the insertion or deletion of several nucleotides not divisible by three from a DNA sequence. This alters the reading frame of the codons downstream of the mutation.
Consequence: It leads to a completely different amino acid sequence from the point of the mutation onwards, often resulting in a premature stop codon (nonsense)
What is the consequence of inserting/deleting 1, 2, or 3 nucleotides?
The consequence of insertions or deletions depends on the number of nucleotides involved:
Insertion or Deletion of 1 or 2 Nucleotides: These types of mutations are frameshift mutations.
They alter the reading frame of the codons downstream of the mutation.
This leads to a completely different amino acid sequence from the point of mutation onward.
Often results in a premature stop codon (nonsense mutation) or a drastically altered, non-functional protein.
Insertion or Deletion of 3 Nucleotides (or any multiple of 3): These are in-frame mutations.
They do not alter the reading frame of the codons.
Instead, they result in the addition or removal of one or more amino acids in the protein.
The impact on protein function can vary; it might be minor or lead to significant functional changes, depending on the role of the added/deleted amino acid(s).
What are the stop codons?
The stop codons (also known as nonsense codons) are specific nucleotide triplets in mRNA that signal the termination of translation. They do not code for any amino acid. The three universal stop codons are:UAA
UAG
UGA (you are away, You are gone, You go away)
What are the biological causes of mutations? (replication errors, slippage during replication)
Biological causes are inherent processes in living organisms that lead to DNA sequence changes. These primarily include replication errors, where DNA polymerase incorporates incorrect nucleotides, and replication slippage in repetitive DNA regions, causing insertions or deletions of repeat units.
What is Huntington’s disease an example of?
Huntington's disease is a genetic disorder resulting from the expansion of CAG trinucleotide repeats in the HTT gene, primarily due to DNA replication slippage.
What is alkylation?
The addition of methyl or ethyl groups to keto
What are two uses of alkylating agents?
Warfare: Such as mustard gas.
Chemotherapy drugs: Used to treat cancer by damaging DNA and preventing cell proliferation.
What is depurination?
What cellular conditions can cause depurination?
Is this common or rare compared to depyrimidation?
Depurination is the hydrolysis of the glycosidic bond between a purine base (adenine or guanine) and deoxyribose, resulting in an apurinic site in the DNA.
Cellular conditions that can cause depurination include:
Low pH: Acidic conditions can accelerate the hydrolysis.
Increased temperature: Higher temperatures promote the reaction.
Presence of reactive oxygen species (ROS): These can damage DNA and lead to depurination.
Depurination is much more common than depyrimidation. It is estimated that thousands of depurinations occur per human cell per day, while spontaneous depyrimidation
What nucleotide is formed when cytosine is deaminated?
When cytosine is deaminated (loses its amino group), it is converted into uracil. Uracil is typically found in RNA but not normally in DNA. In DNA, this conversion creates a mismatch with guanine, as uracil pairs with adenine, not guanine.
What are base analogs? What is an example of a base analog?
Base analogs are modified synthetic bases that can be incorporated into DNA during replication, acting as mutagens because they mispair with natural bases. This leads to point mutations, specifically base substitutions.
An example of a base analog is 5-bromouracil (5-BU). 5-BU is an analog of thymine, but can enolize more readily, allowing it to mispair with guanine instead of adenine, leading to T-A to C-G transitions.
How do intercalating agents cause mutations in DNA? What is an example?
Intercalating agents insert between DNA base pairs, distorting the helix. This confuses DNA polymerase during replication, leading to erroneous nucleotide insertions or deletions, causing frameshift mutations.
Ex: Ethidium Bromide
What nucleotide does 8-oxo-dG base pair with? What cellular conditions cause it to form?
8-oxo-dG (8-Oxo-2'-deoxyguanosine), an oxidized form of guanine, preferentially mispairs with adenine (A) instead of cytosine (C).
Its formation is primarily caused by oxidative stress, which involves the attack of hydroxyl radicals (\text{OH}^\cdot) on guanine residues in DNA.
What causes thymine dimers to form?
Thymine dimers are primarily caused by exposure to ultraviolet (UV) light. UV radiation induces a photochemical reaction where two adjacent thymine bases on the same DNA strand bond covalently, forming a cyclobutane pyrimidine dimer (CPD) or a (6-4) photoproduct.
What are the consequences of the presence of thymine dimers?
Thymine dimers distort the DNA helix, which can block DNA replication and transcription, leading to mutations or cell death if not repaired.
How can thymine dimers be repaired as an example of fixing DNA damage?
Photorepair is a direct repair process where the enzyme DNA photolyase uses energy from visible light to reverse the formation of thymine dimers, restoring the DNA to its original state.
Mutagen
An agent that permanently alters DNA sequences. It can be physical (e.g., UV radiation), chemical (e.g., alkylating agents), or biological (e.g., viruses).