04 - DNA Replication & Repair
1/72
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
73 Terms
All replication systems require:
• Helicases
• Single stranded DNA-binding proteins (SSBs)
• Topoisomerases
• Primase
• DNA Polymerases
• DNA ligase
Initiating replication
requires protein binding to the origin of replication (ori) by sequence recognition and unwinding of DNA.
Replication then proceeds outwards from the origin in both directions
Bacterial chromosomes have ___ ori
1
Initiates at sequences rich in A-T base-pairs. Why?
has less hydrogen bonds, so requires less energy
Replication forks
points where a pair of replicating segments come together
1 replication bubble contains ____ replication forks
2
in prokaryoteas which way does rhe replication fork move
move in opposite directions at ~1000 nucleotides per second until they meet at a point across from the origin
whats the solution to:
The double helix has to be pulled apart to make single - stranded template
hydrogen bonds, a-t rich sequence, initiator proteins and helicase (+ atp hydrolysis)
whats the solution to:
Single stranded DNA has a tendency to fold/reanneal on itself to make hairpins
single stranded binding protein
whats the solution to:
DNA polymerase can’t start from nothing. It needs a 3’OH to build from
dna primase (a dna polymerase)
Single-strand DNAbinding (SSB) proteins
Bind and stabilize single stranded DNA after helicase unwinds DNA the double helix
RNA primer
DNA polymerases cannot initiate DNA synthesis on their own and require a primer on the template to create a 3’OH to build on
what is rna primer synthesized by
DNA primase
DNA primase
a type of RNA polymerase
synthesizes a short (~10 nt) RNA primer which is later removed
whats the solution to:\
When the double helix is opened to make single-stranded DNA, this causes torsional stress and over-winding in front of the replication fork
topiosomerase
whats the solution to:
DNA polymerase has a tendency to fall off the DNA template
sliding clamp
whats the solution to:
As the replication fork progresses, only one of the template strands is oriented for 5’ to 3’ synthesis of the new strand. DNA polymerase CAN’T synthesize in a 3’ to 5’ direction
ozaki fragments, replisome
Topoisomerase I
catalyzes the ‘nicking’ of one DNA strand (allows rotation to relieve strain)
• travels ahead of the fork and relieves this ‘+’ supercoiling
Topoisomerase II
Topoisomerase II catalyzes a double stranded break \Detangles DNA
Role of the sliding clamp
Sliding clamp keeps DNA polymerase on the DNA, but releases it when a double-stranded region is encountered
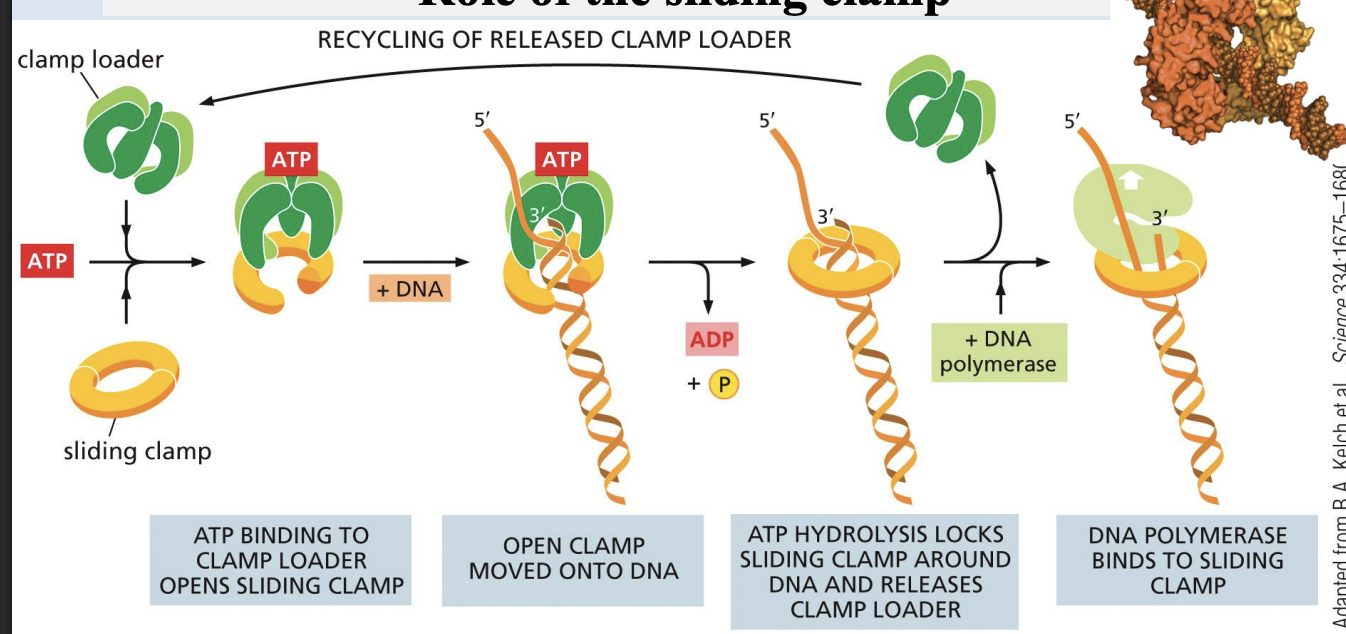
when would a sliding clamp encounter a double-stranded region
when sliding clamp goes through the replication fork and reaches the other strand
leading strand
Polymerization of one strand is towards the replication fork
lagging strand
Polymerization made discontinuously in segments that are each extended away from the fork.
Synthesis of each fragment must wait for the parental strand to expose additional template for primer synthesis (lags slightly behind leading strand synthesis)
Okazaki fragments
Lagging strand is synthesized as small fragments
lagging strand process
Primase adds a RNA primer to initiate each segment
RNA Primer is later removed and gap is filled
DNA ligase covalently connects the Okazaki fragments into a continuous strand
Replisome
A multiprotein complex coordinates the leading and lagging strand
how does the polymerase travel together
Looping of the lagging strand allows the polymerases to travel together.
The loop is released when an Okazaki fragment is completed
The loop forms again for the synthesis of the next Okazaki fragment on the lagging strand template
DNA polymerases require
Template DNA
A primer to which a new nucleotides are added → provides the necessary 3’-OH.
All four deoxyribonucleotides (dATP, dTTP, dGTP, dCTP)
Nucleotide selection is dependent on
base-pairing
DNA polymerase III
extends RNA primers
Which strand has RNA primers?
leading have 1
lagging has multiple
DNA polymerase I
replaces RNA primers.
Where does the 3’OH come from for DNA pol I to build on?
The 3′ OH comes from the end of the adjacent DNA fragment (usually made by DNA polymerase III).
DNA pol III has 3’→ 5’ exonuclease activity. This is important for…?
decrease mutation rate, as it goes back and corrects mistake
DNA pol I has both 3’→ 5’ and 5’ → 3’ exonuclease activity and can degrade DNA or RNA What do you think the 5’ → 3’ exonuclease activity is important for?
important to remove rna primer and replace it
additional challenges of eukaryotic genomes replication
1. Timing with mitosis
2. Nucleosomes
3. Large genome size
4. Chromosome ends
challenge with Timing with mitosis
Eukaryotic DNA replication is coordinated with cell cycle
Bacteria can initiate DNA replication almost continuously
Eukaryotes have a more complex control system regulating/coordinating both DNA replication and progression through the cell cycle
Nucleus/cell divides in M phase (mitosis)
S phase = DNA Synthesis phase (lasts 40 min to 8hr)
Production of histone mRNA also increases dramatically in S phase. Why?
s phase is where dna replication happens
challenge in Nucleosomes in eukaryotic replication
Chromatin remodeling complexes destabilize nucleosomes for DNA replication
Histones are displaced and reestablished behind the replication fork by histone chaperones
The reestablished nucleosomes on each completed strand contain ~ half “old” histones and ~half new. Why is this important?
each strand carries some of the previous histone modifications to pass on epigenetic
challenge in Multiple origins of replication in Eukaryotic cells
Eukaryotic cells replicate their DNA in small portions – called replicons
Each replicon has its own origin of replication. There are ~10,000 to 100,000 different origins in human cells
Initiation of DNA synthesis in a replicon is highly regulated and requires assembly of an ORC (origin of replication complex)
challenge in Replicating the ends of eukaryotic chromosomes
Without intervention, chromosome ends would shorten with each round of division due to a lack of an available 3’OH to complete strand DNA synthesis
Telomerase is responsible for maintaining chromosome ends
Telomerase
Large enzyme composed of RNA and protein subunits
uses reverse transcriptase activity to synthesize DNA
contains its own RNA template
extends the template strand
Primase + DNA polymerase action can then fill in the end of the lagging strand
Telomeres
are the repetitive sequences at the end of chromosomes
Composed of tandem repeats (~1000 x GGGTTA in humans)
Telomeres form a loop that protect the ends of chromosomes
Expression of telomerase is regulated (some cells have much more than others!) Why might some cells need more than others?
Cells that divide frequently need more telomerase
Humans with the genetic condition dyskeratosis congenita have only 1 copy of the telomerase RNA gene. What do you think happens to these individuals?
Humans with only 1 copy of the telomerase RNA gene have reduced telomerase activity → telomeres shorten faster → stem cell exhaustion and tissue failure → symptoms of dyskeratosis congenita.
stress relation to telomeres
Manage stress = manage your telomeres?
What would happen if we treated/manipulated all of our cells to contain high levels of telomerase activity? Any drawbacks?
alot more repeats dna
pretty wasteful
we will get ancancer, this is one of the genes that cancer activates
The low mutation rate in DNA replication results from:
• Accurate selection of nucleotides
• Immediate proofreading → incorrect nucleotides are removed
• Post-replicative repair → repairs DNA damage after replication
• Repair of DNA damage from sources unrelated to replication
environmental damage to dna
DNA is highly susceptible to environmental damage:
• Reactive molecules/chemicals – often cause structural changes
• Thermal energy – usually from normal metabolism → can cause A/G to come off the sugar backbone (depurination)
• Ionizing radiation – causes breaks in DNA backbone
• UV radiation – causes pyrimidine dimers
What is the major source of UV radiation?
sun
Depurination
removes the base from DNA
Deamination
most commonly converts cytosine to uracil
What will happen if DNA replication occurs and this damage has not been corrected?
Unrepaired nucleotide modifications cause mutations!
is depurination common
yes
Researchers use _____ as a read-out for the amount of oxidative damage in cells
8-oxo guanine
Major repair systems / mechanisms:
• Nucleotide excision repair (NER)
• Base excision repair (BER)
• Strand-directed mismatch repair (MMR)
• Homologous recombination
• Nonhomologous end joining (NHEJ)
Nucleotide excision repair (NER)
• Removes bulky lesions (e.g. pyrimidine dimers, chemical alterations)
• Excision nucleases cut the damaged strands on either side of the lesion
• The two strands are separated by DNA helicase so the cut piece is released
• DNA Polymerase fills in the gap using the other strand as a template
• DNA ligase seals the strand
2 distinct pathway of Nucleotide excision repair (NER)
1. Transcription coupled pathway → repairs DNA that is being actively transcribed
2. Global genomic pathway → slower and less efficient → repairs DNA in the remainder of the genome
Why would NER have evolved to prioritize actively transcribed regions?
NER prioritizes actively transcribed regions because damage there immediately threatens gene expression and cell function.
RNA polymerase helps signal where repair is needed, making this process faster and more targeted.
Clinical implications of defects in NER
• Mutations in several different genes involved in NER result in serious consequences. Patients are unable to repair damage caused by exposure to UV light – Xeroderma pigmentosum (XP)
• Must live life protected from all sunlight
• Highly susceptible to developing skin cancer and have a shortened life expectancy
Base excision repair (BER)
Repairs altered base (e.g. 8-oxoguanine, uracil, deaminated 5-methylcytosine)
initiated by DNA glycosylase enzyme that recognizes a specific alteration
DNA glycosylase removes the altered base by cleaving the glycosidic bond attaching the base to deoxyribose suga
AP endonuclease is then required to remove the remaining sugar-phosphate
May have distinct transcription-coupled and global repair pathway
Based on what we’ve covered so far, which of these enzymes would be most useful for detecting and/or repairing a depurination?
A) Muts
B) DNA glycosylase
C) AP endonuclease
D) Excision nuclease
E) DNA helicase
c
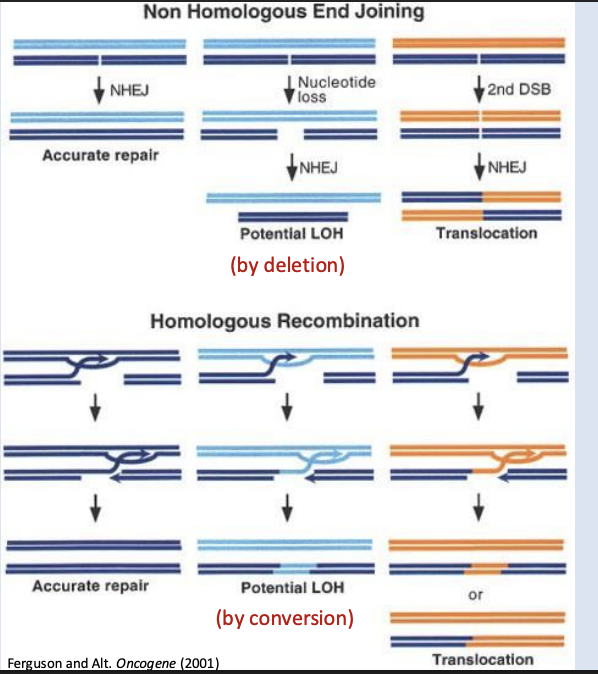
Nonhomologous end joining (NHEJ) - active ness
Highly active pathway in humans for rapid repair
Homologous recombination - active ness
Less active (primarily active during division) pathway for DSB repair
Nonhomologous end joining (NHEJ)
• Ring shaped heterodimeric protein Ku detects DSB and binds both ends
• DSB ends are processed and brought together
• DNA ligase IV seals / joins the strands
• Small deletions are common at the site of repair. Translocations/inversions can also occur
Do small deletions matter?
yes
homologous recombination
• Nuclease digests 5’ ends,
creating overhangs
• RecA/Rad51 catalyzes strand
invasion into another DNA
duplex; Must have homology
(sequence similarity) for
complementary base-pairing!
• Undamaged DNA is used as a
template
• DNA ligase seals the backbone
NHEJ vs Homologous Recombination: Which is more likely to accurately repair the DNA sequence?
Homologous recombination is the more accurate DNA repair mechanism.v
How is homologous recombination possible? What potential sources are there for this homologous sequence in our cells?
Homologous recombination is possible because our cells carry extra copies of the genome (sister chromatids, homologous chromosomes).
The sister chromatid is the preferred and most accurate source during the S and G2 phases of the cell cycle.
What could go wrong in homologous recombination
Loss of heterozygosity (LOH)
Why might homologous recombination cause loss of heterozygosity (LOH)? Could this matter/impact cell function? How? How is this different from the result of homologous recombination during crossing over?
Homologous recombination can cause LOH when it copies information from a homologous chromosome with a different allele.
This can delete or replace a functional gene, especially dangerous if it affects tumor suppressors or essential genes.
This differs from meiotic HR, which increases diversity and doesn’t cause LOH in the same way.
what could go wrong during DSB repair
High
Can create harmful gene fusions or misregulation, associated with cancer