cell mechanics
1/123
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
124 Terms
equation relating energy of a system
ΔU = Q- P ΔV
entropy
ΔS=Q/T
where Q is heat(energy flow)
enthalpy
ΔU = Q-PΔV which can be expanded to a useful equation:
ΔH=Q at constant pressure
Gibbs free energy definition
quantifies if a reaction will occur spontaneously or not
spontaneous or non spontaneous if ΔS<0
non spontaneous
spontaneous or non spontaneous if ΔS>0
spontaneous
spontaneous or non spontaneous if ΔH>0
heat absorbed-non spontaneous
spontaneous or non spontaneous if ΔH<0
heat produced - spontaneous
Gibbs free energy equation
ΔG=ΔH-TΔS
first law of thermodynamics in relation to the cell
ΔU=Q+W
boltzmanns microscopic definition of entropy
S=k ln(Ω)
where Ω is the number of possible configurations of a systemst
ructural requirements of membrane bilayer
semi porous
flexible and deformable
strong enough to withstand external forces and energy fluctuation
surface tension
y = energy/area J/m²
compression energy of bilayer
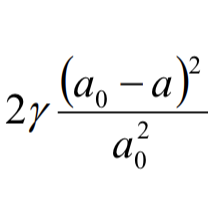
hypotonic
solution concentration is greater inside the cell, osmosis drives water flow into the cell increasing pressure
isotonic
solution inside is equally concentrated to the outside
hypertonic
solution concentration is greater outside the cell, osmosis drives water out
osmotic pressure equation
P=ΔC RT. where C is change in concentration
flicks laws of diffusion(concentration gradient)
J=-D ∂Φ/∂x where Φ is concentration, D=diffusion constant, J=flux
why do molecules move in the presence of thermal energy
it increases collisions which redirects molecules
cytoskeleton components
spectrin
actin
intermediate filaments
microtubules
cytoskeleton polymerisation
cells move by rapid extension and contraction of cytoskeleton fibres driven by polymerisation and depolymerisation
accessory proteins
control filament length
stages of cytoskeleton polymerisation
nucleation
elongation
steady state
nucleation
individual cells associating e
elongation
once the threshold is hit a linear growth phase is entered
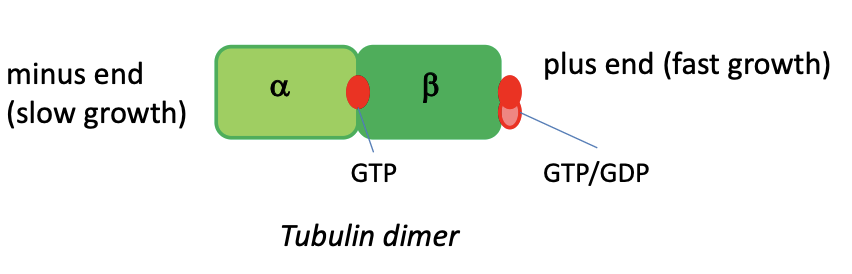
how do the tubular sub units polymerise
alpha and beta tubular sub units involve GTP and GDP (like ATP-ADP) which act as energy converters driving the polymerisation
polymerisation kinetics equation
dn/dt =+KonM-Koff
where Kon/off is the capture/release rate constant, M=concentration of free monomers
Mc(critical concentration)
point at which no net change in filament length
Mc=Koff/Kon
treadmilling
when the growth of the plus end equals the shrinkage at the minus end
properties of polymerisation of filaments
rapid extension and contraction
control of filament length with accessory proteins
chemical to mechanical energy conversion
advantages of polymerisation of filaments
flexibility of response
fmax
(KT/d)ln(M/Mcrit)
f=force
K=boltzmanns constant
T=absolute temp
M=concentration of monomers
Mcrit=Critical concentration
equation illustrating electrons produced by conversion of NADH
NADH—→ NAD^+ + H^+ + 2e^-
equation illustrating electrochemical gradient driving protons back into matrix through ATP synthase
ADP+Pi+4H^+ ——> ATP+H2O+4H^+
ATP synthase
rotates which acts as a source of potential energy
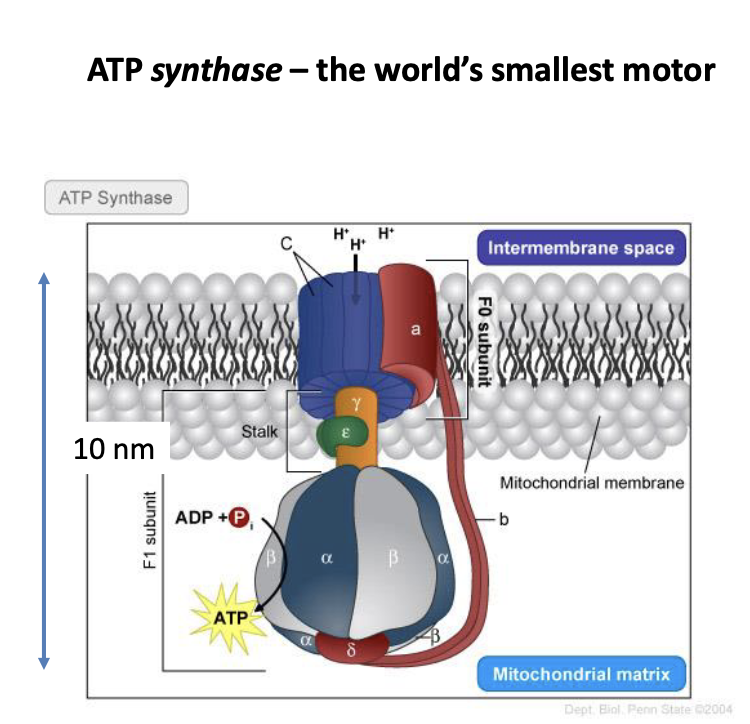
how ATP synthase produces torque
protons flow through due to electrochemical gradient
electrostatic forces between protons and c ring produce rotational motion
ATP synthase energy consumption
1 rotational step of 120° requires 8×10^-20 J
describe how myosin protein crawls along actin filament
myosin binds to filament
ATP capture releases myosin
hydrolysis of ATP to produce ADP leads to shape change and myosin head moves along filament
myosin protein uses energy of one ATP molecule to move 5nm
motor proteins
transport vesicles within the cell
molecule specific binding of vesicle to the motor so cargo is controlled
different types are differentiated due to different behaviour under same force
bidirectional motion of cargo theory
motors will pull cargo in different directions
overall motion is the balance of the forces
motor protein diseases
Alzheimers-loss of nerve cell function is linked to degradation of kinesin and dynein transporting molecules along the axon
glucose—→ carbon dioxide+water
C6H12O6——> CO2+H2O
when is energy extracted in cell processes
stage 1 glycolysis in cell cytoplasm
stage 2 citric acid/Krebs cycle in mitochondria
glycolysis info needed to know for exam
anaerobic
energy loss stage
energy gain stage
energy loss stage reactions
Glucose—→ glucose-6-p using P from ATP (so -1ATP)
|___> produced by two separate reactions:
ATP+H2O—→ ADP+H3PO4
Glucose+H3PO4—> glucose-6-P+H2O
Further oxidation of the carbohydrate:
Glucose-6-P—→ Fructose-6P using P from ATP(so - 1 ATP)
energy gain stage
bisphosphoglycerate*2—→ pyruvate +4ATP (Phosphate from carbohydrate moved to ADP producing 4 ATP total)
citric acid/krebs cycle
driven by enzymes in mitochondrial matrix
preliminary step:
pyruvate—→acetyl-coA+CO2 (turning NAD+ to NADH + H+)citric cycle:
acety-coA+H2O—→Oxaloacetate+CO2 (turning NAD+ to NADH + H+) *3
energy conversion summary
oxidation of c atoms in sugar release high energy electrons
hydrolysis of ATP to ADP and Pi is energetically favourable
cells use enzymes to link energy release from ATP degradation to unfavourable chemical reactions-synthesis, motion
30-36 molecules of ATP produced from 1 glucose, 84 from 1 fatty acid, 40%conversion efficiency
summary energy conversion equations
C6H12O6+6O2+6H2O+36ADP+36P—→6CO2+12H2O+36ATP
NAD+ + 2e- + H+ ←→ NADH
24e- + 6O2 + 24H+ —→ 12H2O
how catalysts aid reactions
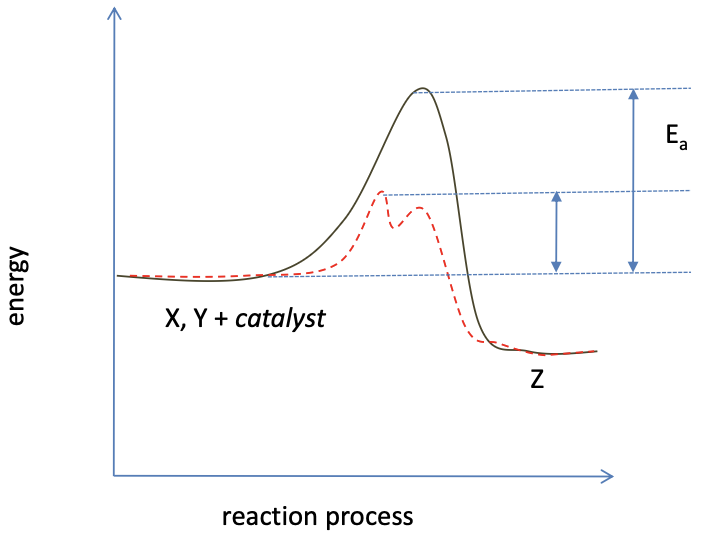
oxidation of glucose
glucose+H3PO4—→ glucose-6-P+H2O (ΔG=+3.2)
why are most reactions energetically unfavourable?
so catalysts control which reactions occur, if spontaneous no control
composition of genes
nucleotide units within a polymer molecule (DNA)
4 DNA nucleotides
Adenine
Guanine
Cytosine
Thymine
nucleotide composition
sugar-phosphate-base
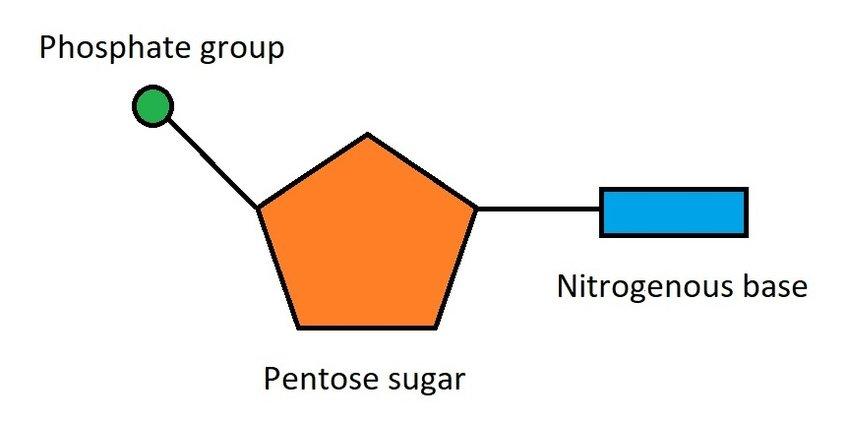
Nucleotide pairing
C&G
A&T
genome
the total DNA content of an organismge
genes
formation of nucleotides form a discrete information set for the production of specific protein molecules
where is DNA stored and why
in chromosomes in the nucleus of each cell,
localises genetic decoding, high molecular concentrations and specialised enzymes
DNA replication
double strand is separated by DNA helicase, driven by ATP hydrolase
matching of base sequence through hydrogen bonding adds deoxyribonucleotide monomers to the polymer strand, directed by DNA polymerase
phosphate groups are added to the backbone
large free energy of the reaction provided by the release of phosphate from ATP-ADP
DNA looping
process where proteins cause a segment of the DNA double helix to bend and form a loop, which brings distant DNA sites close together to regulate cellular processes
energy and entropy considerations of DNA looping
requires energy due to strain on structure
entropy reduction
binding proteins help stabilise despite these constraints
gene expression
translation of genetic code to produce proteins
gene expression stages
transcription- DNA transformed to RNA Uracil replaces Thymine, controlled by ribosome
protein synthesis directed by RNA molecule- translation
transcription
RNA polymerase begins RNA synthesis once promotor sequence is recognised
polymerase moves along DNA synthesising RNA
polymerase encounters terminator sequence
transcription enzyme
RNA polymerase
mechanics of RNA polymerase
once promotor sequence is recognised it clamps to DNA with movable jaws
after transcription of 10 nucleotides a flap closes to form exit tunnel for the synthesised RNA
at the terminator the nucleotide sequence codes a hairpin into the RNA which releases it from DNA
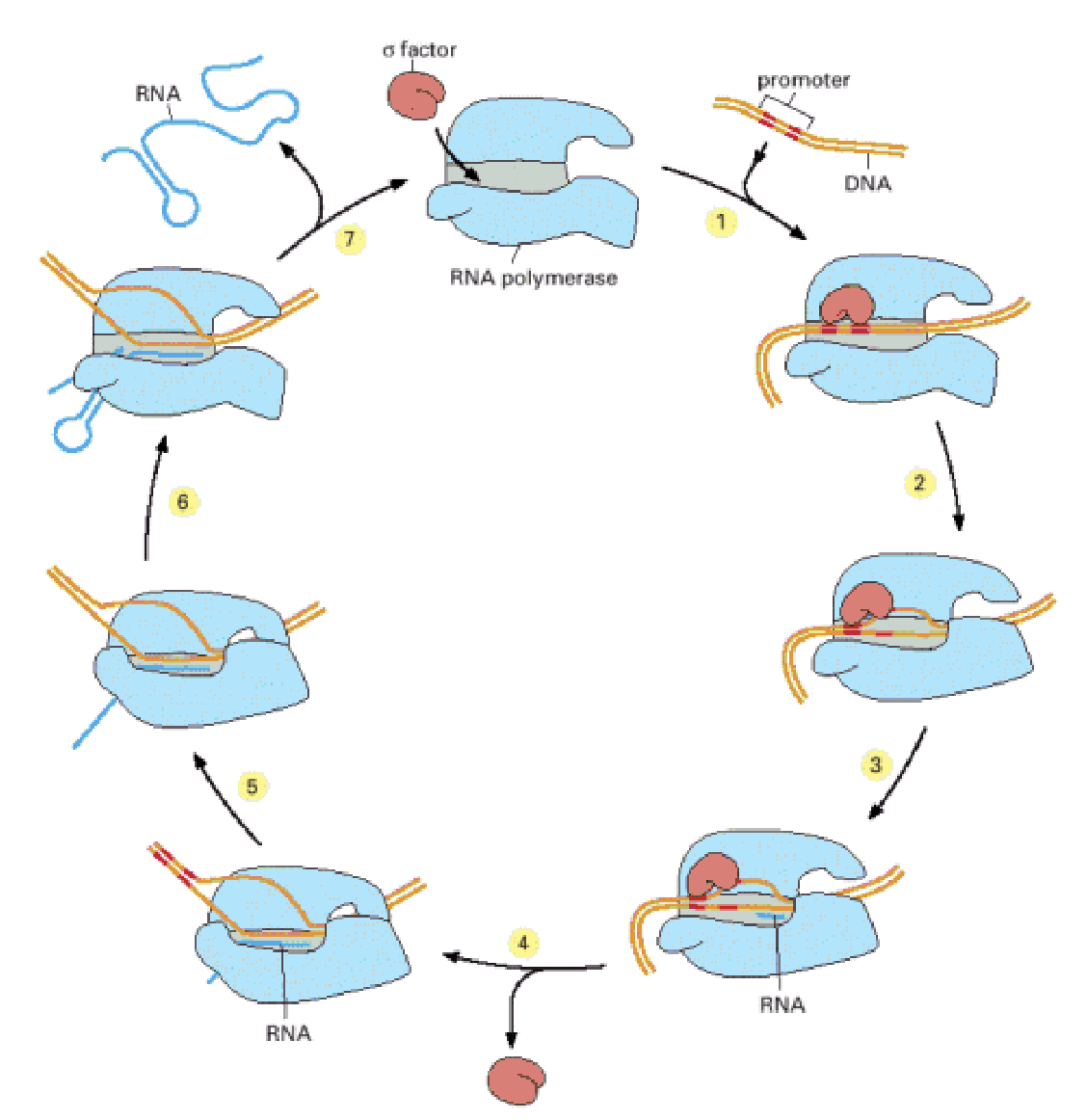
mRNA
completed RNA destined for protein synthesis. they are passed out of nucleus into cytoplasm
codons
3 letter sequences on RNA used to code for specific amino acid, 64 possible sequences-but only 20 amino acids, this built in safety, if a codon is incorrect it may still code for the same protein
example of biological feedback
principles of feedback for system regulation
negative- output I linked back to input in a way that damps the system to reduce output
positive feedback- output linked back to boost system
control system in coli when glucose is scarce
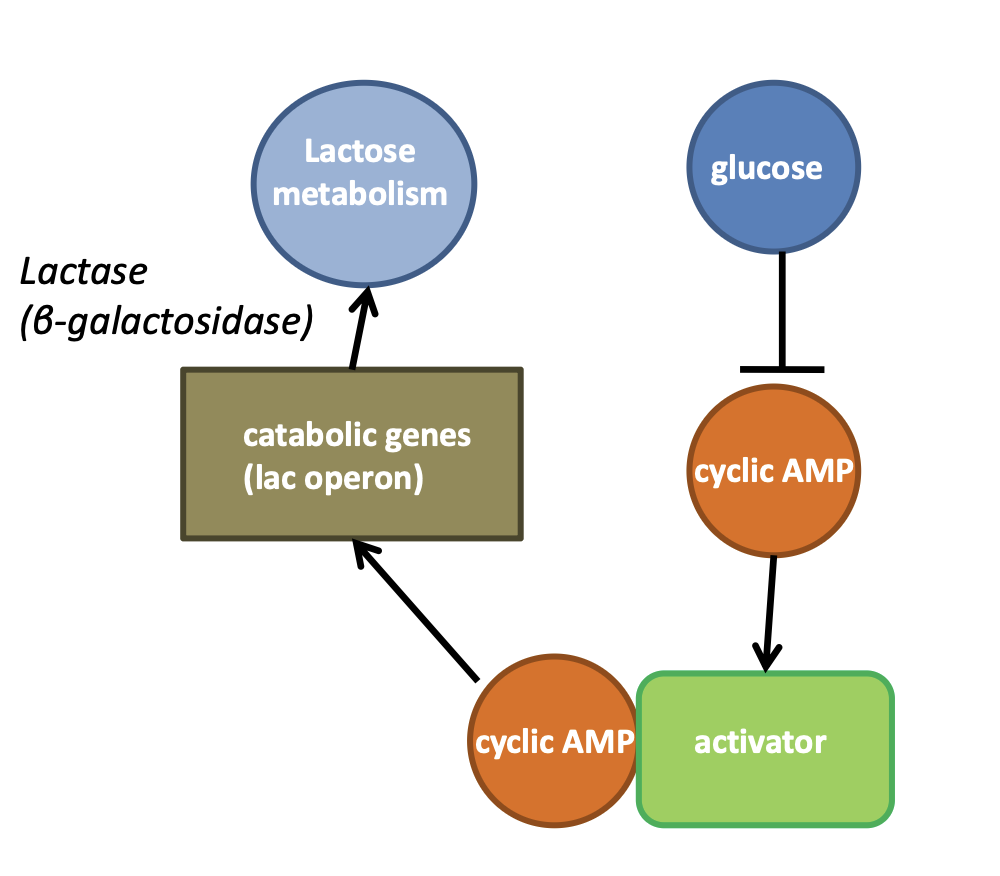
cell signalling
manifests using molecules, signal transmission is via diffusion and cell receptor molecules are the receiver
this triggers a signalling cascade of molecular interactions that manifest A change
membrane based signalling
membrane houses receptors adapted to recognise and bind to ligands (signal molecules)
example of signalling-circadian oscillator
cyclic response to light/darkness. signals production of hormones in pineal gland cortisol and melatonin. the suprachiasmatic nucleus controls this pattern, a part of the brain linked to stimulation of the optic nerve
cell cycle
process of cell growth DNA replication and division
main cycle phases
S phase- DNA replication takes place 8-10 hours
M phase- 2 DNA copies separated into daughter cells 1 hour
in between G1 and 2 phases- cell grows and checks cycle process
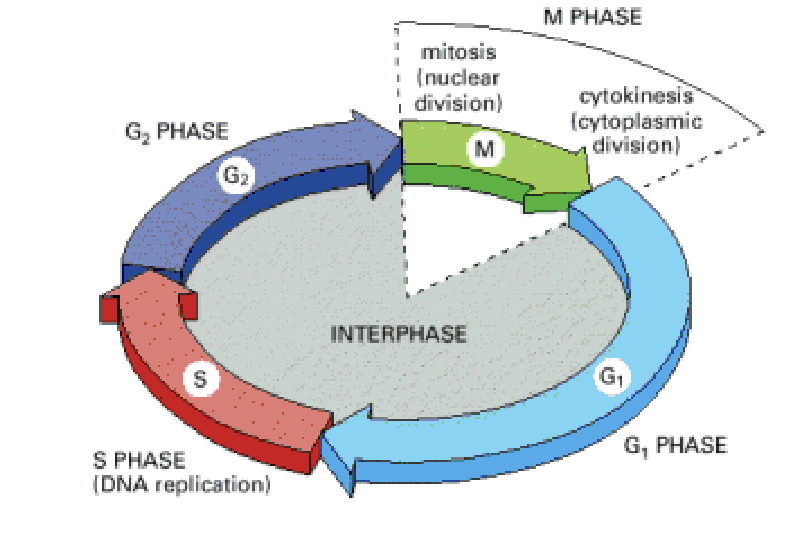
G1 checkpoint
cell control mechanism checks cell status and monitors signals from cellular environment. If cell conditions are not suitable due to chemical cues the cycle is blocked
G2 checkpoint
control mechanism that checks accuracy of DNA copy
cell cycle control system
Timing
synchronicity
directionality-binary on/off switches ensure completion of events
robustness- error checking
flexibility- respond to changes in conditions
does the cycle control system use positive or negative feedback
negative
control based on two families of proteins
Cyclin dependent kinases(cdK)-control major processes of DNA replication, chromosome segregation and mitosis. activity fluctuates, concentration remains constant.
Cyclins- activate Cdk proteins(regulatory molecules) concentration fluctuates.
quality control is initiated at what checkpoints
DNA replication checkpoint- ensure faithful reproduction of genes
Spindle attachment checkpoint- ensure complete attachment of chromosomes
DNA damage checkpoints- ensure damaged genes are not copied
Apoptosis
if damaged cells cannot be repaired they enter a series of molecular interactions which lead to death
activation of p53 by the DNA damage checkpoint
Mdm2 and p53 molecules phosphorylated
Mdm2 releases from p53 and becomes stable
p53 molecule initiates transcription of cdk inhibitor protein-halts cell cycle
transcription of activator proteins release cytochrome C from mitochondria
steps of apoptosis-an example of positive feedback
caspases cut up the cell into small pieces
macrophages (white blood cells) ingest pieces
stages of mitosis(6)
prophase
prometaphase
metaphase
anaphase
telophase
cytokinesis
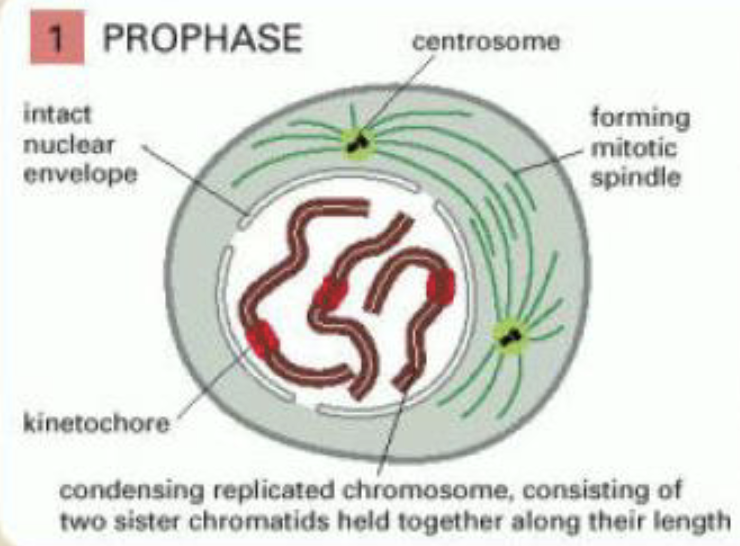
prophase
replicated chromosomes condense
outside the nucleus the mitotic spindle assembles between two centrosomes which have replicated and moved apart
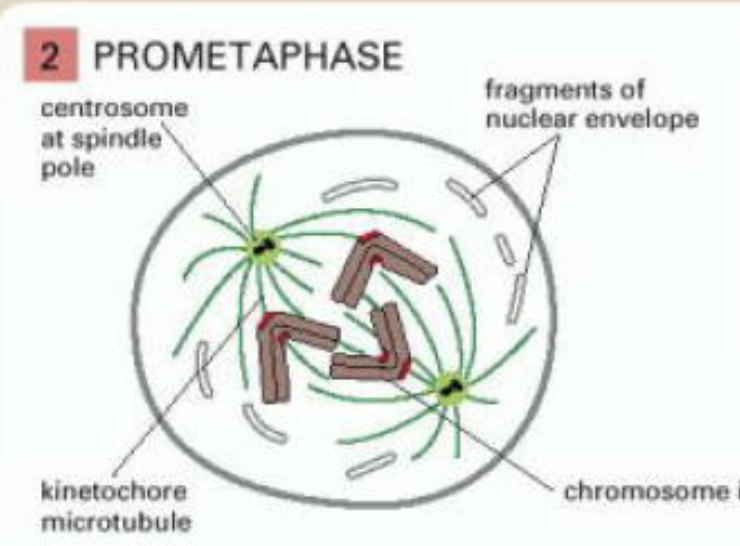
prometaphase
begins with breakdown of nuclear envolope
chromosomes attach to spindle microtubules via kinetochores
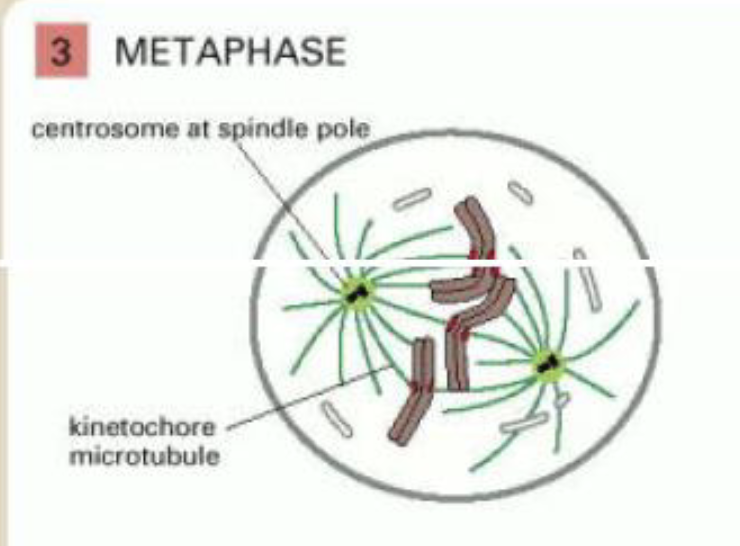
metaphase
chromosomes aligned at equator of the spindle, midway between spindle poles
kinetochore microtubules attach sister chromatids to opposite poles of spindle
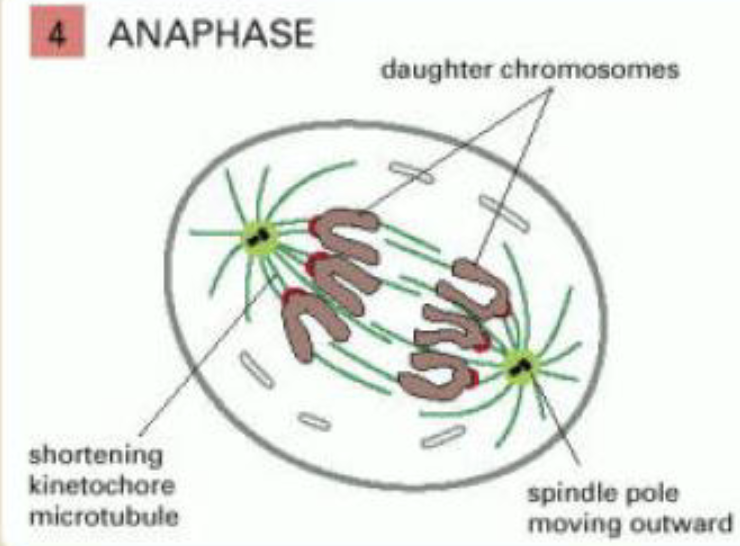
anaphase
sister chromatids synchronously separate to form two daughter chromosomes, pulled in polar direction
kinetochore microtubules get shorter and spindle poles move apart
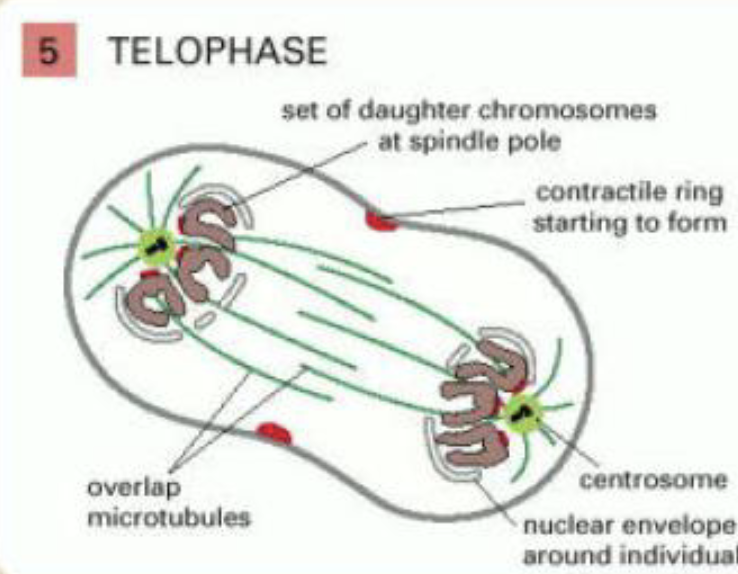
telophase
two sets of daughter chromosomes at poles of spindle decondense
new nuclear envelope reassembles around each set
division of cytoplasm begins with assembly of contractile ring
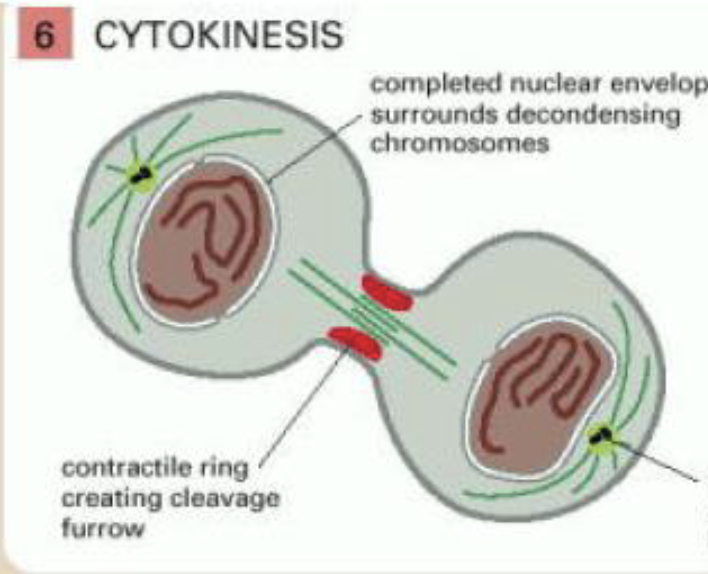
cytokinesis
cytoplasm is divided in two by contractile ring of actin and myosin filaments, pinching the cell
internal vesicles recruited to the cleavage furrow to provide additional membrane required to form the gained surface area of daughter cells
example of anti-cancer drug
paclitaxel inhibits microtubule disassembly so prevents formation of mitotic spindle, cells stall in G2 and enter apoptosis
simple equation describing thee proliferation of cells over time
N=2^t/TD
where TD = cell cycle time
t= time
exponential growth equation
dN/dt=rN
where N=number of cells
r=rate constantlo
logistic growth equation
dN/dt=rN(1-N/K)
K=carrying capacity
what proteins trigger cell proliferation
growth factor proteins
survival factors
proteins secreted by neighbouring cells, they attach to cell surface receptor molecules which down regulates the apoptotic pathway