BIO 205 Exam 2
0.0(0)
0.0(0)
Card Sorting
1/99
Earn XP
Study Analytics
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
100 Terms
1
New cards
Deoxyribonucelic Acid (DNA)
Stores genetic information and is replicated using proteins
2
New cards
RNA world hypothesis
proposes time in evolution where RNA both stored genetic information and catalyzed its own replication
3
New cards
Nucleic Acid
polymer of nucleotide monomers
4
New cards
3 components of nucleotide
1. Phosphate group
2. Five- carbon sugar
3. Nitrogenous (nitrogen-containing) base
5
New cards
Ribonucleotides
monomers of DNA
* Have ribose as their sugar
* Has an -OH group (bonded to 2’ carbon)
* Have ribose as their sugar
* Has an -OH group (bonded to 2’ carbon)
6
New cards
Deoxyribonucleotides
monomers of DNA
* Sugar is deoxyribose (=lacking oxygen)
* Both sugars have -OH group bonded to 3’ carbon
* Involved in nucleotide bonding (phosphodiester linkage)
* Sugar is deoxyribose (=lacking oxygen)
* Both sugars have -OH group bonded to 3’ carbon
* Involved in nucleotide bonding (phosphodiester linkage)
7
New cards
Purines
contain nine atoms in their one ring:
* Adenine (A)
* Guanine (G)
* Adenine (A)
* Guanine (G)
8
New cards
Pyrimidines
Contain six atoms in their one ring:
* Cytosine (C)
* Uracil (U)- only in RNA
* Thymine (T)- Only in DNA
“CUT of Py”
* Cytosine (C)
* Uracil (U)- only in RNA
* Thymine (T)- Only in DNA
“CUT of Py”
9
New cards
Condensation Reactions
* How Nucleic acids polymerize
* Creates bonds (removes H+ and OH-/H2O and replaces them with new bonds)
* Creates bonds (removes H+ and OH-/H2O and replaces them with new bonds)
10
New cards
Phosphodiester lineage
Join ribonucleotides together
* Phosphate group on 5’ carbon of one nucleotide
* -OH group on 3’ carbon of another
* Polymer produced is RNA/DNA
* Phosphate group on 5’ carbon of one nucleotide
* -OH group on 3’ carbon of another
* Polymer produced is RNA/DNA
11
New cards
Sugar-Phosphate backbone
* directional (5’ - 3’ direction)
* one end has unlinked 5’ phosphate group
* one end has unlinked 3’ hydroxyl group
* one end has unlinked 5’ phosphate group
* one end has unlinked 3’ hydroxyl group
12
New cards
Nucleic Acid polymerization
* Can take place in cells by use of enzymes
* Potential energy raised → 2 additional phosphate group
* Creates nucleotide triphosphates “activated nucleotides”
* ex: Adenosine Triphosphate (ATP)
* Potential energy raised → 2 additional phosphate group
* Creates nucleotide triphosphates “activated nucleotides”
* ex: Adenosine Triphosphate (ATP)
13
New cards
X-ray cyrstallography
used to measure distances between atoms in DNA
* predicted helical structure
* predicted helical structure
14
New cards
Primary Structure of DNA
written by listing sequences of bases by single-letter abbreviations
* ex: 5’ -ATTAGC- 3’
* ex: 5’ -ATTAGC- 3’
15
New cards
Secondary Structure of DNA
* DNA polymerized → phosphate groups
* sugar-phosphate backbone
* equal number of purines and pyrimidines
* sugar-phosphate backbone
* equal number of purines and pyrimidines
16
New cards
Grooves
attachment sites for enzymes of transcription and replication
17
New cards
Major Groove
backbones closer
18
New cards
Minor Groove
backbones farther
19
New cards
complementary and antiparallel
DNA strands in a double helix are
20
New cards
Tertiary Structure of DNA
* DNA forms compact three dimensional structures in cells
* Compaction
* 2 forms of DNA tertiary structure
* 1. DNA wraps around histones
* DNA twists to form supercoils
* Chromatin form
\
* Compaction
* 2 forms of DNA tertiary structure
* 1. DNA wraps around histones
* DNA twists to form supercoils
* Chromatin form
\
21
New cards
Histones
Compact DNA strands and impact chromatin regulation
22
New cards
Supercoils
DNA twists to form
* Have to be untwisted to replicate or create proteins
* Have to be untwisted to replicate or create proteins
23
New cards
Chromatin
* material of which the chromosomes of organisms other than bacteria are composed
* supercoiled DNA and is relatively spread out
* supercoiled DNA and is relatively spread out
24
New cards
Euchromatin
loosely packed DNA
25
New cards
Heterochromatin
highly a compacted DNA molecule which only occurs during cell division
26
New cards
Nucleosome
DNA wrapped 8 histones
27
New cards
Arginine and lysine
DNA histones that are positively charged
28
New cards
Phosphates
DNA is negatively charged
29
New cards
Biological reservoir of information
* __***Stores information***__ required for organism’s:
* growth
* reproduction
* Consists of __***sequences of nucleotides***__ in nucleic acid
* 4 nitrogenous bases
* __***Sequence***__ determines everything
* Sequence order is important
* growth
* reproduction
* Consists of __***sequences of nucleotides***__ in nucleic acid
* 4 nitrogenous bases
* __***Sequence***__ determines everything
* Sequence order is important
30
New cards
DNA Replication
1. 2 strands separated → hydrogen bonds
2. Free deoxynucleotides form hydrogen bonds → ==complementary bases of template strand==
3. Complementary base pairing allows each strand to be copied exactly → ==identical daughter molecules==
31
New cards
Complementary Strand
Phosphodiester linkages form to create new strand
32
New cards
Double Helix Structure of DNA
held together by:
* phosphodiester linkages (backbone)
* hydrogen bonds (bases)
* Hydrophobic interactions (helical structure)
\
* phosphodiester linkages (backbone)
* hydrogen bonds (bases)
* Hydrophobic interactions (helical structure)
\
33
New cards
Functional Group roles in DNA
* makes molecule __***stable and resistant***__ to __**degradation**__
* Stability of DNA key to effectiveness of reliable ==information-storage molecule==
* Stability of DNA key to effectiveness of reliable ==information-storage molecule==
34
New cards
Primary Structure of RNA
* Four Types of nitrogenous bases extending from sugar
* phosphate backbone
1. RNA contains ribose (instead of deoxyribose)
2. RNA contains uracil (instead of thymine)
* Both bind to adenine
3. 2’ -oh group on ribose is ==more reactive== than -H
4. RNA is much ==less stable==
\
* phosphate backbone
1. RNA contains ribose (instead of deoxyribose)
2. RNA contains uracil (instead of thymine)
* Both bind to adenine
3. 2’ -oh group on ribose is ==more reactive== than -H
4. RNA is much ==less stable==
\
35
New cards
Secondary Structure of RNA
* results from complementary base pairing: A → U; G → C
* Bases typically form **hydrogen bonds** with complementary bases on same strand
* RNA strand folds over, forming h**airpin structure:**
* Bases on one part fold over and align with bases on other part
* Two sugar-phosphate strands are **antiparallel**
* Bases typically form **hydrogen bonds** with complementary bases on same strand
* RNA strand folds over, forming h**airpin structure:**
* Bases on one part fold over and align with bases on other part
* Two sugar-phosphate strands are **antiparallel**
36
New cards
Tertiary Structure of RNA
* Forms when secondary structures fold into more complex shapes
* RNA shapes are more diverse in shape, size and reactivity than DNA
* Can have ==enzymatic function==
* RNA shapes are more diverse in shape, size and reactivity than DNA
* Can have ==enzymatic function==
37
New cards
DNA Polymerase
* Enzymes that catalyzes DNA synthesis
* Several types
* Works in one direction
* Add deoxyribonucleotides only in 3’ end of a growing DNA chain
* DNA synthesis always proceeds in 5’ → 3’ direction
* Several types
* Works in one direction
* Add deoxyribonucleotides only in 3’ end of a growing DNA chain
* DNA synthesis always proceeds in 5’ → 3’ direction
38
New cards
Endergonic
DNA polymerization requires input of energy
39
New cards
Deoxyribonucleoside triphosphates (dNTPs)
* Have 3 phosphates
* high potential energy b/c of their closely packed __***phosphate groups***__
* have enough potential energy to make formation of __***phosphodiester bonds exergonic***__
* high potential energy b/c of their closely packed __***phosphate groups***__
* have enough potential energy to make formation of __***phosphodiester bonds exergonic***__
40
New cards
Replication Bubble
an unwound and open region of a DNA helix where DNA replication occurs
* Forms when DNA is being synthesized
* Origin of replication:
* Bacteria → one and form only one
* Eukaryotic cell have many on each chromosome
\
* Forms when DNA is being synthesized
* Origin of replication:
* Bacteria → one and form only one
* Eukaryotic cell have many on each chromosome
\
41
New cards
Replication Forks
Structure that forms within the long helical DNA during DNA replication.
* Synthesis is __**bidirectional**__
* replication bubbles grow in __**2 directions**__
* Synthesis is __**bidirectional**__
* replication bubbles grow in __**2 directions**__
42
New cards
DNA helicase
Highly conserved group of enzymes that unwind DNA
* Breaks hydrogen bonds between two DNA strands to separate them
* Single-strand DNA-binding proteins (SSBPs) attach to separated strands to prevent closing
* Breaks hydrogen bonds between two DNA strands to separate them
* Single-strand DNA-binding proteins (SSBPs) attach to separated strands to prevent closing
43
New cards
Topoisomerase
* makes single-stranded cuts in DNA
* cut and pass double stranded DNA
* cut and pass double stranded DNA
44
New cards
Primer
short stretches of DNA that targets unique sequences and help identify a unique part of genome
* Serve as a starting point for DNA synthesis
* 10 to 25 nucleotides long
* Serve as a starting point for DNA synthesis
* 10 to 25 nucleotides long
45
New cards
Lagging Strand
* discontinuous strand
* Strand synthesized away from replication fork
* Occurs because DNA synthesis must proceed in 5’ → 3’ direction
* Strand synthesized away from replication fork
* Occurs because DNA synthesis must proceed in 5’ → 3’ direction
46
New cards
Discontinuous replication hypothesis
* Primase synthesizes new RNA primers on lagging strand as replication fork opens
* DNA polymerase synthesizes short fragments of DNA along lagging strand
* Fragments are linked into continuous strand
* DNA polymerase synthesizes short fragments of DNA along lagging strand
* Fragments are linked into continuous strand
47
New cards
Okazaki Fragments
short sections of DNA formed at the time of discontinuous synthesis of lagging strand during replication of DNA
48
New cards
Telomeres
region at the end of eukaryotic chromosome
* do not contain genes
* do not contain genes
49
New cards
Telomere Replication
1. 3’ end of lagging strand forms single-stranded “overhang”
2. Telomerase binds to overhand → uses RNA as template for DNA synthesis
3. As telomerase move down the new strand, it adds more short DNA sequences at the end of parent strand
4. Once overhang is long enough, normal DNA synthesis can occur
50
New cards
Exonuclease active site
* Mismatched deoxyribonucleotide moves to site where it does fit
* Site catalyzes removal of incorrect deoxyribonucleotide
* Site catalyzes removal of incorrect deoxyribonucleotide
51
New cards
Mismatch repair
occurs when mismatched bases are corrected after DNA synthesis is complete
52
New cards
Nucleotide excision repair
* similar details to the mismatch repair above
* protein complex *recognizes* kink
* *removes* damaged single-stranded DNA
* Uses intact strand as *template* for new DNA
* **DNA** **ligase** *links* repair strand to original undamaged DNA
* protein complex *recognizes* kink
* *removes* damaged single-stranded DNA
* Uses intact strand as *template* for new DNA
* **DNA** **ligase** *links* repair strand to original undamaged DNA
53
New cards
Gene Expression
the process of converting information in DNA into functioning molecules within the cell
54
New cards
Central Dogma
DNA → RNA → Proteins
* ==Genes== → stretches of DNA that code for proteins
* **DNA sequence** codes for RNA sequence
* **RNA sequence** codes for sequence of amino acids in protein
* ==Genes== → stretches of DNA that code for proteins
* **DNA sequence** codes for RNA sequence
* **RNA sequence** codes for sequence of amino acids in protein
55
New cards
Transcription
process of using DNA template to make complementary RNA
* making a copy of information
* making a copy of information
56
New cards
Translation
process of using information in mRNA to synthesize proteins
* interprets nucleotide “language” to amino acids
* interprets nucleotide “language” to amino acids
57
New cards
Reverse transcriptase
Some viruses never go through a DNA stage
* Synthesizes DNA from an RNA template
* Gene flow is RNA → DNA
* Synthesizes DNA from an RNA template
* Gene flow is RNA → DNA
58
New cards
Genetic Code
specifies how a sequence of nucleotides codes for a sequence of amino acids
59
New cards
Codon
* mRNA
* Group of three bases that specifies particular amino acid
* Distinguish from DNA triplets
* Group of three bases that specifies particular amino acid
* Distinguish from DNA triplets
60
New cards
Redundant code
All but two amino acids are encoded by more than one codon
61
New cards
Unambiguous Code
One codon never codes for more than one amino acid
62
New cards
Non- overlapping Code
Codons are read one at a time
63
New cards
Universal Code
All codons specify the same amino acids in all organisms (with minor exceptions)
64
New cards
Conservative Code
If several codons specify the same amino acid, the first two bases are usually identical
65
New cards
Mutation
any permanent change in an organism’s DNA:
* Modification in cell’s information archive
* New alleles
* Change in its genotype
* Modification in cell’s information archive
* New alleles
* Change in its genotype
66
New cards
Point Mutation
mutations result from one or a small number of base changes
67
New cards
Chromosome- level Mutation
* mutations are larger in scale
* may change chromosome number → polyploidy or aneuploidy → or structure
* may change chromosome number → polyploidy or aneuploidy → or structure
68
New cards
Missense Mutation
Point Mutation
* mutations change an **amino acid** in protein
* mutations change an **amino acid** in protein
69
New cards
Silent Mutation
Point Mutation
* mutations do *not change amino acid sequence* due to **redundancy** in the code
* mutations do *not change amino acid sequence* due to **redundancy** in the code
70
New cards
Frameshift Mutation
Point Mutation
* mutations shift reading frame, altering meaning of **all** subsequent codons
* mutations shift reading frame, altering meaning of **all** subsequent codons
71
New cards
Nonsense Mutation
Point Mutation
* mutations change codon that specifies an amino acid into **stop codon**
* mutations change codon that specifies an amino acid into **stop codon**
72
New cards
Inversion Mutation
Chromosome Mutation
* segment of chromosome breaks off, flips around, and rejoins
* segment of chromosome breaks off, flips around, and rejoins
73
New cards
Translocation Mutation
Chromosome Mutation
* section of chromosome breaks off and becomes attached to another chromosome
* section of chromosome breaks off and becomes attached to another chromosome
74
New cards
Deletion Mutation
Chromosome Mutation
* segment of a chromosome is lost
* segment of a chromosome is lost
75
New cards
Duplication Mutation
Chromosome Mutation
* segment of chromosome in present in multiple copies
* segment of chromosome in present in multiple copies
76
New cards
Karyotype
complete set of chromosome in cell
77
New cards
Genetic Screen
Method of identifying organisms with mutations in genes that produce a product of interest
* Assumption: mutate an enzyme, **you will not get the molecules** produced by the enzyme
* Assumption: mutate an enzyme, **you will not get the molecules** produced by the enzyme
78
New cards
RNA Polymerase
synthesizes an RNA version of the instructions stored in DNA
* Uses ribonucleoside triphosphates (NTPs)
* Matches complementary bases to one strand of DNA
* perform template-directed synthesis in **5’ → 3’ direction**
* does not require a primer to begin transcription
* Cannot initiate transcription on its own
\
* Uses ribonucleoside triphosphates (NTPs)
* Matches complementary bases to one strand of DNA
* perform template-directed synthesis in **5’ → 3’ direction**
* does not require a primer to begin transcription
* Cannot initiate transcription on its own
\
79
New cards
Bacterial Promoters
40-50 base pair long and contains **two sites** which are recognized by sigma
* -10 box and -35 box
* -10 box and -35 box
80
New cards
Termination
occurs when RNA polymerase transcribes a signal
* Codes for RNA that forms a **hairpin structure**
* Causes the RNA polymerase to separate from the RNA transcript
* Codes for RNA that forms a **hairpin structure**
* Causes the RNA polymerase to separate from the RNA transcript
81
New cards
Elongation
RNA polymerase reads the DNA template:
* Nucleotides are added to the 3’ end of the RNA
* Nucleotides are added to the 3’ end of the RNA
82
New cards
Transcription in Eukaryotes
* 3 RNA polymerase
* Larger promoters, including TATA Box
* General transcription factors
* At termination: poly (A) signal is transcribed (rather than a hairpin)
* Larger promoters, including TATA Box
* General transcription factors
* At termination: poly (A) signal is transcribed (rather than a hairpin)
83
New cards
Poly A signal Sequence
The end of the transcript
* Transcribed in Eukaryotes during termination
* Transcribed in Eukaryotes during termination
84
New cards
snRNPs
Small nuclear ribonucleoproteins
85
New cards
RNA Splicing
Form of RNA processing in Eukaryotes:
Allows different mRNAs and proteins to be produced from a single gene
* Cut out: introns
* Kept: exons
* Catalyzed by snRNPs
Allows different mRNAs and proteins to be produced from a single gene
* Cut out: introns
* Kept: exons
* Catalyzed by snRNPs
86
New cards
4 Steps of RNA Splicing
1. __snRNPs__ bind to 5’ exon -intron and 3’ intron - exon boundaries and to an A near the end of intron
2. Other snRNPs join complex to form a **spliceosome**
3. The intron forms a single- stranded __stem plus a loop__ (lariat)
4. The lariat is cut out and 2 exons are linked. Intron → degraded
87
New cards
Caps and Tails
Pre- mRNA are processed by 2 events:
1. ==5’ cap==: modified guanine nucleotide **enables ribosomes to bind and protect from degradation**
2. ==Poly (A) tail:== 100-250 adenine nucleotides **needed for translation and protect from degradation**
3. Product → mature mRNA
1. ==5’ cap==: modified guanine nucleotide **enables ribosomes to bind and protect from degradation**
2. ==Poly (A) tail:== 100-250 adenine nucleotides **needed for translation and protect from degradation**
3. Product → mature mRNA
88
New cards
Template
* read by RNA polymerase
* mRNA will have a **complementary sequence** to this strand
* Does **NOT carry the “code” to** create a protein
* mRNA will have a **complementary sequence** to this strand
* Does **NOT carry the “code” to** create a protein
89
New cards
Non-template
* not read by RNA polymerase
* Complementary to template
* mRNA will have **same sequence**
* **Carries the “code”** for making a protein
* Complementary to template
* mRNA will have **same sequence**
* **Carries the “code”** for making a protein
90
New cards
Transcription and Translation in Eukaryotes
* mRNAs are **synthesized and processed** in nucleus
* **Mature** mRNAs are transported to cytoplasm for translation by ribosomes and polyribosomes form
* **Mature** mRNAs are transported to cytoplasm for translation by ribosomes and polyribosomes form
91
New cards
Transfer RNA (tRNA)
Adapter molecule used in translation
* Ex: aminoacyl tRNA → linked to its amino acid
* Ex: aminoacyl tRNA → linked to its amino acid
92
New cards
Structure of tRNA
* short: 75 - 95 nucleotides long
* form secondary structures by folding into a **stem-and-loop**
* form secondary structures by folding into a **stem-and-loop**
93
New cards
Aminoacyl-tRNA synthetase
“charge” the tRNA using ATP
* Catalyze the addition of amino acids to tRNA
* Catalyze the addition of amino acids to tRNA
94
New cards
Wobble Pairing
hydrogen-bonded pairing between two nucleotides generally occurring between two RNA molecules that does not follow the Watson-Crick base pair rules
* The anticodon’s third position can form a nonstandard base pair
* The anticodon’s third position can form a nonstandard base pair
95
New cards
A site
Acceptor or aminoacyl → tRNA carries an amino acid
* remains if there is a codon-anticodon match
* remains if there is a codon-anticodon match
96
New cards
P site
Peptidyl → holds growing peptide chain
* peptide bond forms between the amino acid on the A-site tRNA and the polypeptide on the ___ tRNA
* the ribosome moves down the mRNA by one codon and all three tRNAs move down one position
\
* peptide bond forms between the amino acid on the A-site tRNA and the polypeptide on the ___ tRNA
* the ribosome moves down the mRNA by one codon and all three tRNAs move down one position
\
97
New cards
E site
Exit → tRNAs without amino acids exit the ribosome
\
\
98
New cards
Initiation in Bacteria
* begins near the AUG start codon
* small ribosomal subunit binds to the ribosome binding site → Shine-Dalgarno sequence
* 3 step process:
1. mRNA binds to a small ribosomal subunit
2. Initiator tRNA bearing f-Met binds to the start codon
3. Large ribosomal subunit binds so that the initiator tRNA is in the P site
* small ribosomal subunit binds to the ribosome binding site → Shine-Dalgarno sequence
* 3 step process:
1. mRNA binds to a small ribosomal subunit
2. Initiator tRNA bearing f-Met binds to the start codon
3. Large ribosomal subunit binds so that the initiator tRNA is in the P site
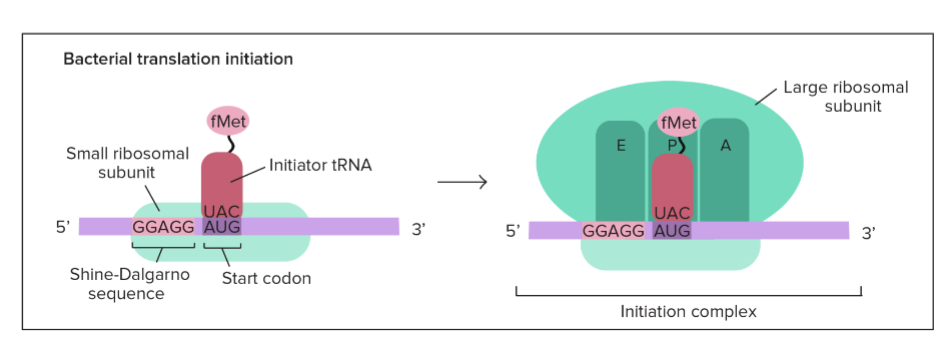
99
New cards
Initiator tRNA
first tRNA in initiating translation
* carries a modified methionine (f-Met) in bacteria
* carries a modified methionine (f-Met) in bacteria
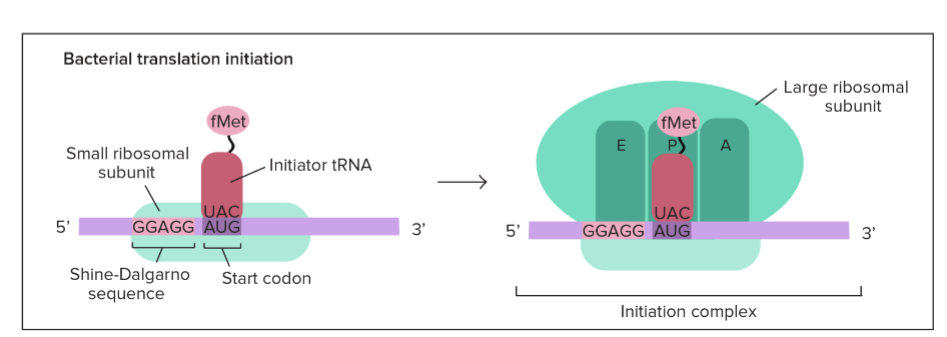
100
New cards
Initiation in Eukaryotes