DNA Damage & Repair
1/58
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
59 Terms
Phenotype
Observable characteristics of a person, an organ, or a cell
Genotype
combination of alleles that a person possesses at a single locus (or at a number of loci)
Disease phenotype
specific manifestations that arise in response to the differential expression of just one or a small number of genes that may be harmful
Character or trait
Observable manifestations that are not disease-associated
Multiple characters/traits make up a phenotype
Ex. blue eyes; blood group O
Genetic variation
Changes in the base sequence of our DNA → change in phenotype
Environmental factors and epigenetic effects contribute to the ___
Development of disease or change in phenotype
Epigenetics
Addition of chemical moieties to DNA (ex. methylation, acetylation), leading to a particular gene to be silenced or activated
Change in phenotype can happen even w/o change of DNA sequence through epigenetic modification
Mutation
Process that produces altered DNA sequences AND the outcome of that change
Both a verb and a noun: process of making the change and the outcome of the change
Mutation → DNA variants (alternative forms of DNA) → >1% polymorphisms, <1% rare variants
Depending on how prevalent the specific mutation is worldwide, it’s called single nucleotide variant (less common; <1%) or single nucleotide polymorphism (more common; >1%)
4 consequences of Mutations
Normal phenotype (ex. height)
Disease phenotype
No obvious effect on the phenotype
Very rarely, some beneficial effect
Mutations originate as a result of changes in our DNA that are __
Not corrected by cellular DNA repair systems
DNA changes are occasionally induced by radiation and chemicals in our environment, but the great majority arise from ___
endogenous sources
Types of Genetic Variation
No change in DNA content
Change in DNA content → Net loss/gain of DNA sequence
Genetic Variation: No change in DNA content
Ex. SNPs: one nucleotide is replaced w/ another nucleotide
Most common DNA changes are on a small scale and involve only a single nucleotide or a very small number of nucleotides
Small scale change (point mutations) often have no obvious effect on the phenotype (silent/neutral mutations)
Single Nucleotide Polymorphisms (SNPs)
Change in a single nucleotide
Most common type of genetic variation in the human genome
SNP variation accounts for a lot of physical variations like blood type
Account for ~75% of DNA changes
Estimated that there is 1 SNP/1000 base pairs in the human genome
~3 million SNPs/person
Genetic Variation: Change in DNA content; Net loss/gain of DNA sequence
Ex. Trisomy, deletion
Change in the copy number of whole nuclear DNA molecules are almost always harmful
Most are embryologically lethal except: Down syndrome, 21 trisomy; Edward’s syndrome, 18 trisomy; Turner Syndrome, X; Kleinfelter’s syndrome, X
Many result in spontaneous abortion and some give rise to developmental disorders
ABO Blood Group
Gene inactivation in normal individuals
Most genetic variation has a neutral effect on the phenotype, but a small fraction is harmful
Immune system genes
Polymorphic: undergo somatic rearrangements to produce different variants
Genes involved in identifying microbial pathogens → Constant positive selection to maximize diversity in the proteins involved in antigen recognition
Variation typically happen d/t 3 major events
Recombination
Independent Assortment
Various mutational events
Variation: Recombination
Prophase I of meiosis I: homologous chromosomes exchange of chromosomal segments
More common in the subtelomeric regions
Important for producing the variation that happens at the sperm or egg stage → No two eggs or sperm are the same
Variation: Independent Assortment (of paternal and maternal homologs)
Metaphase I: random lining up and separating, movement of 1 chromosome is not dependent on another one (completely random)
Variation: Various mutational events
Endogenous chemical damage to DNA
Chemical damage to DNA caused by external mutagens
DNA replication errors
Chromosome segregation and recombination errors
3 Types of Endogenous Chemical Damage to DNA
Hydrolytic damage
Oxidative damage from normal cellular metabolism
Aberrant DNA methylation
Hydrolytic damage
Disrupt covalent bonds that hold bases to sugars, cleaving the base from the sugar to produce an abasic site → loss of purine bases (depurination)
Abasic site: location in a DNA or RNA molecule where a nucleotide has lost its nitrogenous base
Hydrolysis: use of water to break a bond
Sometimes break bonds that are not supposed to be broken leading to hydrolytic damage
Oxidative Damage
Most significant are superoxide anions (O2–), hydrogen peroxide (H2O2), and hydroxyl radicals (OH)
Too much ROS → DNA strand break
Aberrant DNA methylation
Many cytosines in our DNA are methylated by methyltransferases
Cells also use S-adenosyl methionine (SAM) as a methyl donor in a non-enzymatic reaction to methylate different types of molecules
Sometimes SAM can inappropriately methylate DNA to produce harmful bases → unwanted gene silencing
3 Types of External Mutagens
UV radiation (sunlight): covalent bonding b/w pyrimidines
Bind to the same strand instead of the other strand → unwanted linkage
High energy irradiation (X-Rays): Generate ROS → DNA strand break
Mutagenic chemicals (ex. cigarette smoke, automobile fumes) → bulky DNA adducts inserting itself b/w DNA strands → obstruction and distortion of the double helix
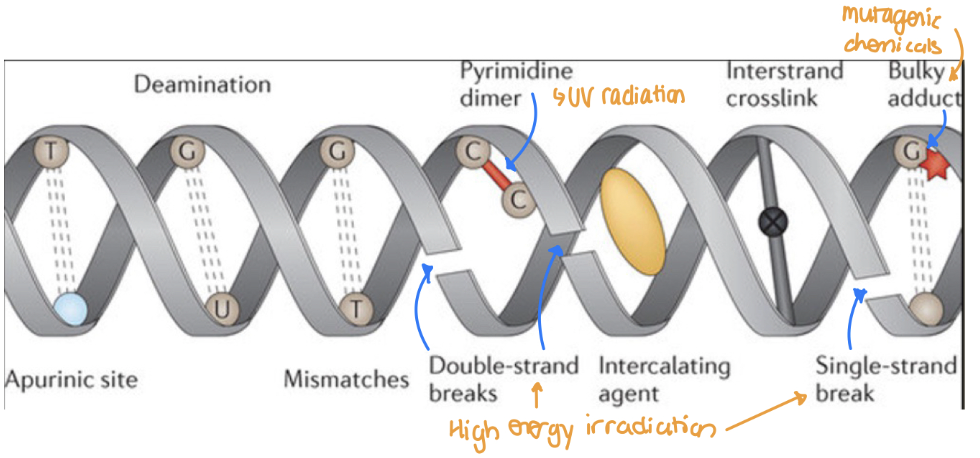
Types of DNA Repair
Single stranded break repair:
Base excision repair (BER)
Nucleotides excision repair (NER)
Mismatch repair (MMR)
Double stranded break repair:
Non-homologous end joining (NHEJ)
Homologous recombination (HR)
Base excision repair (BER)
Repairs lesions where a single base has either been modified or excised by hydrolysis to leave an abasic site
Only can fix small lesions (1 base), it is fast and precise, and basically like spell check coming in to replace 1 mistyped letter
Available throughout the cell cycle
Base excision repair (BER) Process
(1) To replace a modified base by the correct one: specific DNA glycosylase cleaves the sugar–base bond to delete the base, producing an abasic site
(2) Endonuclease and phosphodiesterase remove residual sugar-phosphate from the abasic site
Endonuclease clears the area
Phosphodiesterase cleaves the phosphodiester bond
(3) DNA polymerase fills gap
(4) DNA ligase seals
DNA mismatch repair (MMR)
Repairs erroneous insertion, deletion, mis-incorporation of bases during DNA replication and recombination
Like you wrote a sentence poorly and deleted it to rewrite it → fixing typos
More important in S phase but not restricted to only S phase
DNA mismatch repair (MMR) Process
(1) Mismatch recognized on the daughter strand
(2) Identifies which one was the good strand and which was the wrong one → acts during/after replication
(3) Mismatch and surrounding nucleotides fully excised, creating a large gap
Fold an area with error and then clears it
(4) DNA polymerase fills gap
(5) DNA ligase seals
Nucleotide excision repair (NER)
Repair bulky, helix-distorting DNA lesions (UV induced T-dimers)
Like a sentence riddled with so many spelling mistakes due to someone smashing your keyboard that you just delete the chunk and rewrite it
More important in G1 phase but not restricted to only G1 phase
Nucleotide excision repair (NER) Process
(1) Lesion detected and damage site is opened
(2) DNA cleaved some distance away on either side of the lesion, generating an oligonucleotide of about 30 nucleotides containing the damaged site
(3) Damaged oligonucleotide discarded
(4) DNA polymerase fills the gap, using the template strand as a guide
(5) DNA ligase seals
Homologous recombination (HR)-mediated DNA repair
Highly accurate repair mechanism → requires a homologous intact DNA strand to be available to act as a template strand
Operates in S and G2 phase (before mitosis), using a DNA strand from the undamaged sister chromatid as a template to guide repair
Homologous recombination (HR)-mediated DNA repair Process
(1) Double stranded DNA break
(2) Proteins come together and cleave the area
(3) Overhang falls close to the homologous strand (template) and use that to fill the sequenced area (creates a loop-like structure and then extends the strand using the homologous chromosome as a template)
(4) Goes back to original strand
(5) Polymerase fills the gap
(6) Ligase seals it
Non homologous end joining (NHEJ)
No template strand needed → broken ends are fused together
Always available to cells
Does not have the requirement of a template strand which only is available after DNA replication (S and G2 phase)
Most important for the repair in G1 phase, before the DNA replication
Xeroderma Pigmentosum
Inability to repair damage caused by UV light (NER)
Xeroderma Pigmentosum Mechanism
Defective NER → Unrepaired UV-induced mutations → Xeroderma Pigmentosum → predisposition to skin cancer
Malfunction of nucleotide excision repair (NER) → thymine dimers remain → block replication → unrepaired UV-induced mutations → Xeroderma Pigmentosum → predisposition to skin cancer
Inheritance of Xeroderma Pigmentosum
autosomal recessive
Clinical Features of Xeroderma Pigmentosum
Dry skin, pigment accumulation (ex. freckles), clouding of cornea, keratitis (inflammation of cornea), cancer of the eyelets or conjunctiva, etc.
Cannot repair the damage caused by sunlight → sensitive to sunlight → produce lots of melanin and predisposed to cancer
Before children turn 10 years old, typically have a diagnosis of cancer
Typically skin cancer (ex. basal cell carcinoma, squamous cell carcinoma, malignant melanoma) d/t sensitivity of UV rays
30% of kids w/ the disease can also have severe progressive neurological dysfunction (ex. seizures, cognitive disability, difficulty speaking, difficulty hearing, movement disorder, etc.)
Onset of Xeroderma Pigmentosum
Typically diagnosed ~2years old
Signs appear in infancy or early childhood
Xeroderma pigmentosum with vs. without neurological dysfunction
With: live to ~20 years old (longevity shortens)
Without: live to ~30 years old
Severe Combined Immunodeficiency (SCID)
Deficiency in both B and T lymphocyte functions (both antibody and cell mediated immunity are gone)
Inheritance of SCID
X-linked recessive or autosomal recessive
Lab Findings in SCID
low IgG, IgA, and IgE levels
Most common form of SCID
Mutations in the gene encoding the common gamma chain (γc), a protein that is shared by receptors for interleukins (ILR2 receptor)
ILR2 receptor detects signals from the outside and sends messages to the inside
Not functioning → can’t activate cytotoxic T cells or B cells → immune cells malfunction → no cell-mediated or antibody-mediated immunity → cannot fight infections
Most severe form of SCID
Defects in non-homologous DNA end joining (NHEJ) mechanism (double-strand break repair)
Treatment of SCID
W/o bone marrow transplant, they can’t survive the first 2 years of life
If the bone marrow transplant happens before 3 months of age, longevity increases significantly
Clinical manifestations of SCID
“BE SURE”
Bone abnormalities
Ear infections (8+ years)
Sinus infections (2+ years)
Unexplained failure to thrive
Recurrent pneumonia
Excessive time on antibiotics
Hereditary Non-polyposis Colorectal Cancer (HNPCC) aka Lynch Syndrome
Most common form of hereditary colorectal cancer
Cause of HNPCC aka Lynch Syndrome
mutations in genes involved in DNA mismatch repair (MMR proteins)
Inheritance of HNPCC aka Lynch Syndrome
Autosomal dominant
Clinical Features of HNPCC aka Lynch Syndrome
Propensity to develop right-sided, flat adenomas at a young age
Develop adenomas (glandular benign tumors) at the same rate as individuals in the general population; however they are more likely to progress to adenocarcinoma (cancer)
50-70% risk of developing colorectal cancer and other cancers (ex. stomach, hepatobiliary, small intestine, urinary tract, ovarian, endometrial)
Treatment of HNPCC aka Lynch Syndrome
High dose aspirin (300mg) reduces the progression from adenoma to adenocarcinoma but it is not clinically practiced d/t risk of bleeding
Prevalence of HNPCC aka Lynch Syndrome
1/300 Americans
Bloom Syndrome
Mutation in BLM gene → defective ReQ helicase → defective unwinding of DNA → defective homology mediated DNA repair → Bloom syndrome
BLM gene codes for ReQ helicase
ReQ helicase participates in the unwinding of DNA in the DNA replication process and is also required for the homology mediated DNA repair mechanism
Inheritance of Bloom Syndrome
Autosomal recessive
Clinical features of Bloom Syndrome
Short stature (<5ft)
Butterfly-shaped rash
High pitched voice
Long, narrow face
Small lower jaw
Prominent nose and ears
People w/ Bloom Syndrome live until their ___
20-30s
Rare and lethal disorder d/t high risk for cancer (ex. Skin, colorectal, leukemia, lymphomas)