MCB Ch. 7: Microbial Growth
1/126
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
127 Terms
eukaryotic reproduction
(fungi)
haploid or diploid
asexual or sexual (microorganisms)
prokaryotic reproduction
haploid only (1 chromosome)
asexual
prokaryotic replication methods
binary fission
budding
filamentous
binary fission
one cell grows and divides into two identical cells
time of this process can differ
bigger size/higher complexity → slower replication
budding
yeast and bacteria
one mother cell gives rise to multiple buds
filamentous
attached and hair-like
ex: streptomyces - source of antibiotics
chromosome replication and partitioning
origin of replication (oriC)
terminus (ter)
chromosome must replicate before replication continues (rate-limiting step) ???
septum forms → cell divides
origin of replication (oriC)
within DNA
site where replication starts
terminus (ter)
site where replication ends
chromosome partitioning
Replisome pushes, or leads to condensation of, daughter chromosomes to opposite ends
this system exists in almost 70% of 400 sequenced bacterial genomes
ParA/ParB proteins
ParA/ParB
proteins in C. crescentus
C. crescentus floats around and attaches to hard surface (stalk) when it is ready to replicate
ParA
protein ~ polymerase
polymerize and binds to ParB
as it depolymerizes, it pulls the other chromosome to the right (away from the stalk)
ParB
sequence-specific protein ~ binding
binds to parS (DNA sequence)
ParB binds to pole close to stalk and parS, holding chromosome to left side
Z ring formation
role in septation
several steps
1. selection of site for septum formation
2. assembly of Z ring
3. assembly of cell wall synthesizing machinery
4. constriction of cell and septum formation
what is the Z ring made of
FtsZ
where does the Z ring form?
in the absence of MinCDE (center of cell)
what type of membrane protein is the Z ring?
peripheral protein (not attached to membrane)
what happens when the Z ring depolymerizes?
separation and septum of the two cells form
FtsZ polymers no longer holds Z ring together → 2 daughter cells separate
assembly of cell wall synthesizing machinery
peptidoglycan synthesizing enzymes
anchoring proteins
peptidoglycan synthesizing enzymes
main function is to prevent cell from growing too much and exploding
building more peptidoglycan allows for bacterial growth
anchoring proteins
FtsA and ZipA help attach FtsZ to membrane
constriction of cell and septum formation
MinCDE system
polymerize at one pole and then depolymerize and re-polymerize at another pole
goes left and right at all times
dictates location
inhibits function of Z ring
penicillin binding proteins (PBPs)
synthesizes and maintains peptidoglycan (bacterial cell wall)
targeted by β-lactam antibiotics, like penicillin, which disrupt peptidoglycan synthesis, leading to bacterial cell death.
transpeptidases (TPases)
type of PBP -
catalyzes the cross-linking between peptide chains in the peptidoglycan
they form the peptide bridges that stabilize the bacterial cell wall by linking the 3rd AA on one NAM-pentapeptide to the 4th AA (D-Ala) on another NAM
final step in cell wall synthesis
first step of peptidoglycan synthesis → cellular growth
1. UDP-NAM-pentapeptide is synthesized in cytoplasm
second step of peptidoglycan synthesis → cellular growth
2. bactoprenol (lipid carrier facing the cytoplasm) + UDP-NAM-pentapeptide → lipid I
- joined via pyrophosphate bond
- bacteria uses UTP instead of ATP to activate sugar
- UDP has 2 phosphates
third step of peptidoglycan synthesis → cellular growth
3. UDP-NAG (activated NAG) + lipid I → lipid II
fourth step of peptidoglycan synthesis → cellular growth
4. bactoprenol flips (flippase) lipid II across cytoplasmic membrane (in/cytoplasm → out/periplasmic space), bringing NAM/NAG (peptidoglycan building blocks) to periplasmic space to be added to peptidoglycan
- enzymes come in to move NAM/NAG
fifth step of peptidoglycan synthesis → cellular growth
bactoprenol is flipped back to cytoplasm
autolysins
enzyme breaks down peptidoglycan to allow for growth
- NAM and NAG adds where it is broken
goal of antibiotics
make peptidoglycan weak enough for cell to lyse
- penicillin
- vancomycin
- bacitracin
penicillin
Binds to transpeptidase and prevents bridge between 3rd AA to 4th AA → cell wall is weak
vancomycin
Doesn't allow removal of terminal alanine
bacitracin
Inhibits bactoprenol from flipping from the outside back inside, can't bring NAM/NAG again
E. coli vs S. aureus in peptidoglycan
E. coli:
- DAP
- gram-negatives have a direct link between 3rd and 4th AA
S. aureus:
- L-Lys
- gram-positives have a bridge between 3rd and 4th AA
- has a 5th lysine
Both:
- secondary amino group which allows for 3 peptide bonds
lysosomes can break
cell walls
why can't lysosomes break pseudomurein
bc of the β-1,3 glycosidic bond
vibroid cell wall growth and cell shape
FtsZ, MreB, CreS, CrvA
when division occurs, there is expression of FtsZ and formation of Z ring
CrvA
gives vibrio shape in V. cholerae, V. parahaemolyticus, V. vulnificus
V. fischeri is not a human pathogen (glows in squid)
not a good antibacterial target
spherical bacteria
e.g., S. aureus
elongation:
no elongation
division:
FtsZ forms Z ring to initiate division
rod bacteria
e.g., B. subtillis and E. coli
elongation:
MreB elongates bacteria into rod shape
division:
FtsZ forms Z ring
MreB continues elongation
vibroid bacteria
e.g., C. crescentus
elongation:
MreB elongates
crescentin localizes to inner curve and inhibits growth, causing bending
division:
FtsZ initiates division
MreB elongates
crescentin provides curvature
growth
increase in cellular constituents
growth leads to
increase in cell number (divide)
discontinuous
increase in cell size
continuous, though temporary - it eventually divides
what does growth refer to
population growth rather than growth of individual cells
growth curve
plotted as logarithm number of viable cells vs. time
5 distinct phases:
- lag, log/exponential, stationary, death, long-term stationary
viable cells
can replicate
metabolically active
1. lag phase
exists for a short or long period of time, or is skipped
cell is preparing to grow/replicate
transcribing and translating peptidoglycan building enzymes (FtsZ, MinCDE, ZipA, FtsA)
when does lag phase occur?
From stationary or death phase into a fresh medium
From exponential phase into a fresh medium of different chemical composition
- cell has to adapt to new medium
- biphasic growth (glucose → lactose)
From a rich to a poor culture (medium shift)
- learning how to make stuff they were previously given for free
biphasic growth
2 phases
ex: media has both lactose and glucose
uses glucose first because it's a monosaccharide (lactose is a disaccharide)
- if you can use lactose, you can use glucose (since lactose = glucose + galactose), but a cell that uses glucose may not be able to use lactose
when does lag phase not occur?
Cells growing exponentially into a fresh medium of the same chemical composition
- ex: lactose → glucose (bc lactose has glucose)
2. log/exponential phase
Replicating as fast as possible
growing rate is maximal
- healthiest
- prokaryotes grow faster than eukaryotes (because they are smaller, simpler, and haploid)
- smaller cells grow faster than larger cells
population is most uniform
can be physically seen
3. stationary phase
Running out of nutrients and space, collects toxic material
population growth eventually ceases
- limit reached
- accumulation of waste (bacteria stops replicating when environment changes)
total number of viable cells remains constant, no net change
no building enzymes (FtsZ)
4. death phase
Exponential but not as fast as log
two alternative hypotheses
1. viable but not culturable (VBNC)
2. programmed cell death (PCD)
viable but not culturable (VBNC)
includes E. coli (HUS), V. cholerae (cholera), and L. monocytogenes (listeriosis)
cannot replicate on a petri dish
decreased in size and slowed metabolism, "sleeping beauty"
programmed cell death (PCD)
apoptosis
cell is no longer viable and kills itself to release content into environment for a neighboring cell to absorb
5. long-term stationary phase
evolution (strongest survives)
small percent may mutate and thrive in waste material
bacterial population continually evolves → natural selection (surviving mutant)
Mutations can lasts months to years
'Birth' and 'death' rates are balanced
Growth advantage in stationary-phase (GASP) phenotype
measurement of growth rate and generation time
only for binary fission during exponential growth (not budding because it produces more than 2)
if a number is not given, N0 = 1
generation time
(g)
varies from minutes to hours to days
growth rate constant
(k)
generations per hour
larger number = faster growth
ex:
60/10 mins = 6 generations
60/20 = 3 g
60/60 = 1 g
n
generation number
Nt
total final cell number
Nt = 2^n
.
solving for populations reproducing by binary fission
Nt = N0 x 2^n
solving for n (number of generations)
n = log Nt - log N0 / 0.301
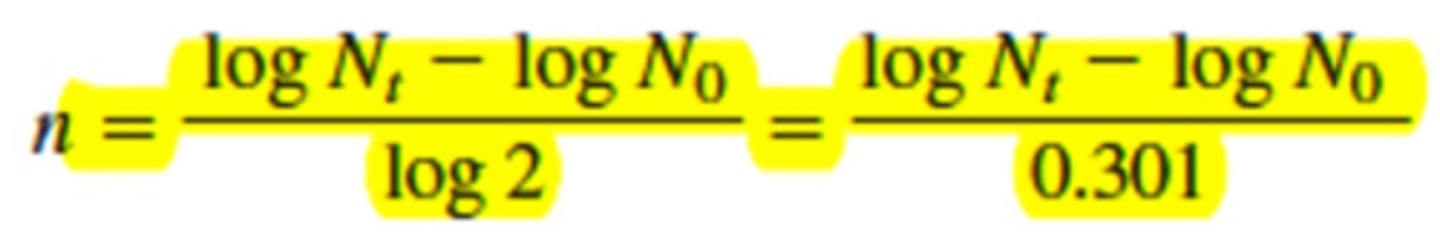
solving for k (growth rate constant)
k = log Nt - log N0 / 0.301(t)
solving generation doubling time
Nt = 2N0
k=1/g
or g=1/k
what conditions are organisms grown in
fairly moderate environmental conditions
few places on earth are considered sterile
extremophiles (archaea)
- non-pathogenic because the environment they thrive in is not something humans have
microbes are adapting
what environment do microbes grow in
hypotonic (water goes in)
they have developed to prevent water from rushing in
reduce osmotic concentration
mechanosensitive (MS) channels ~ hypotonic
contractile vacuoles in protists (eukaryote)
mechanosensitive (MS) channels
pressure on membrane makes it tighter and causes places to open for water to exit
if the channels are closed, water cant get out and cell explodes
as they feel the pull on the membrane, they open
- tighter: channel opens
- cell gets smaller: channel closes
contractile vacuoles in protists
vacuoles push water out
Increase internal solute concentration
compatible solutes (in cytoplasm) ~ hypertonic (from increase in salt)
osmophiles
compatible solutes (in cytoplasm)
Equate inside and outside solute concentration
By increasing inside solute concentration
Cannot inhibit enzymatic activities
Prolene: can increase conc of prolene which increases solute conc to be equal to outside, no net loss
Must be increasing concentration of something that does not cause harm to them
Helps protect since they lack peptidoglycan
Nonhalophile
Not salt lovers
Halotolerant
tolerates salt
- S. aureus
extreme halophiles
requires salt
- Halobacterium spp.
(archaea ~~ extremophile)
compatible solutes
helps bacteria/cells to survive in high solute concentration (salt in environment)
- must equate or else water will go outside
osmotolerant
tolerate stuff outside but doesn't prefer to grow in it
higher solute → lower water activity (aw)
less water → aw decreases
- water availability necessary for microbial growth
all domains of life
Acidophiles
(0-5.5)
archaea
Neutrophiles
(5.5-8)
bacteria and protists
Alkaliphiles/alkalophiles
(8-11.5)
marine microorganisms
most microbes maintain an internal pH near
neutrality
- dies if internal pH is < 5.5-5.0 → denaturation
acid tolerance response
pumps protons out
- too many protons → pH drops → cell death
some synthesize acid and heat shock proteins that refold and protect proteins against denaturation of proteins
many microorganisms change what?
the pH of their habitat
microbes cannot regulate what?
their internal temperature
cardinal growth temperatures
minimum, optimum, maximum
30-40º C difference between minimum and maximum
- No bacteria grows at 0 and 100º C
optimum temperature
Optimum is generally closer to maximum
Bc heat is energy, enzymatic activity increases as temp increases (which is at an optimum temp, which is the best speed)
Anything higher denatures the protein
Psychrophiles
cold lovers
Psychrotolerant
Tolerate cold
Mesophiles
moderate temperature loving (human, body temp)
majority of pathogens are ___?____ though one is psychrotolerant
mesophiles
psychrotolerant - Listeria monocytogenes
Thermophiles
heat loving
Hyperthermophiles
extreme thermophiles, grow in extreme heat
fight against denaturation
psychrophiles
protein structure:
more α-helix, less β-sheets
β-sheets can freeze and come together which is what they want to avoid
more polar, less hydrophobic amino acids
fatty acid chains:
more unsaturated (more double bonds) and shorter
Fatty acids found in bacteria and isoprenes (euk)
Compatible solutes to decrease freezing point
hyperthermophiles
protein structure:
presence of chaperones & HSP
more H bonds and proline
fatty acid chains:
more saturated, branched and higher molecular weight
most are certain species of archaea
monolayers and ether linkages
histone-like proteins stabilize DNA27
Histones are in eukaryotes; histone-like found outside of eukaryotes
thiglycolate test tubes
as it solidifies, it inhibits oxygen from getting into the bottom → makes a gradient/mostly found at top
what color is the oxic zone when oxygen is present
pink
obligate aerobe
growth only at top
+SOD
+catalase
+peroxidase