MMSC 490 Exam 1
1/127
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
128 Terms
peroxisomes
-organelles of plant and animal cells
-site of many oxidative reactions
cell wall
-organelle of bacteria and plants
-rigid layer composed of carbohydrates and proteins
-porous and easily penetrated
plasma membrane
-organelle of bacteria, plant and animal cells
-bilayer of phospholipids and associated proteins
-provides separation from inside of cell and external environment
nucleoid
-organelle of bacteria
-contains 1 circular molecule of DNA
-not surrounded by membrane
ribosomes
-organelle of bacteria, plant and animal cells
-site of protein synthesis
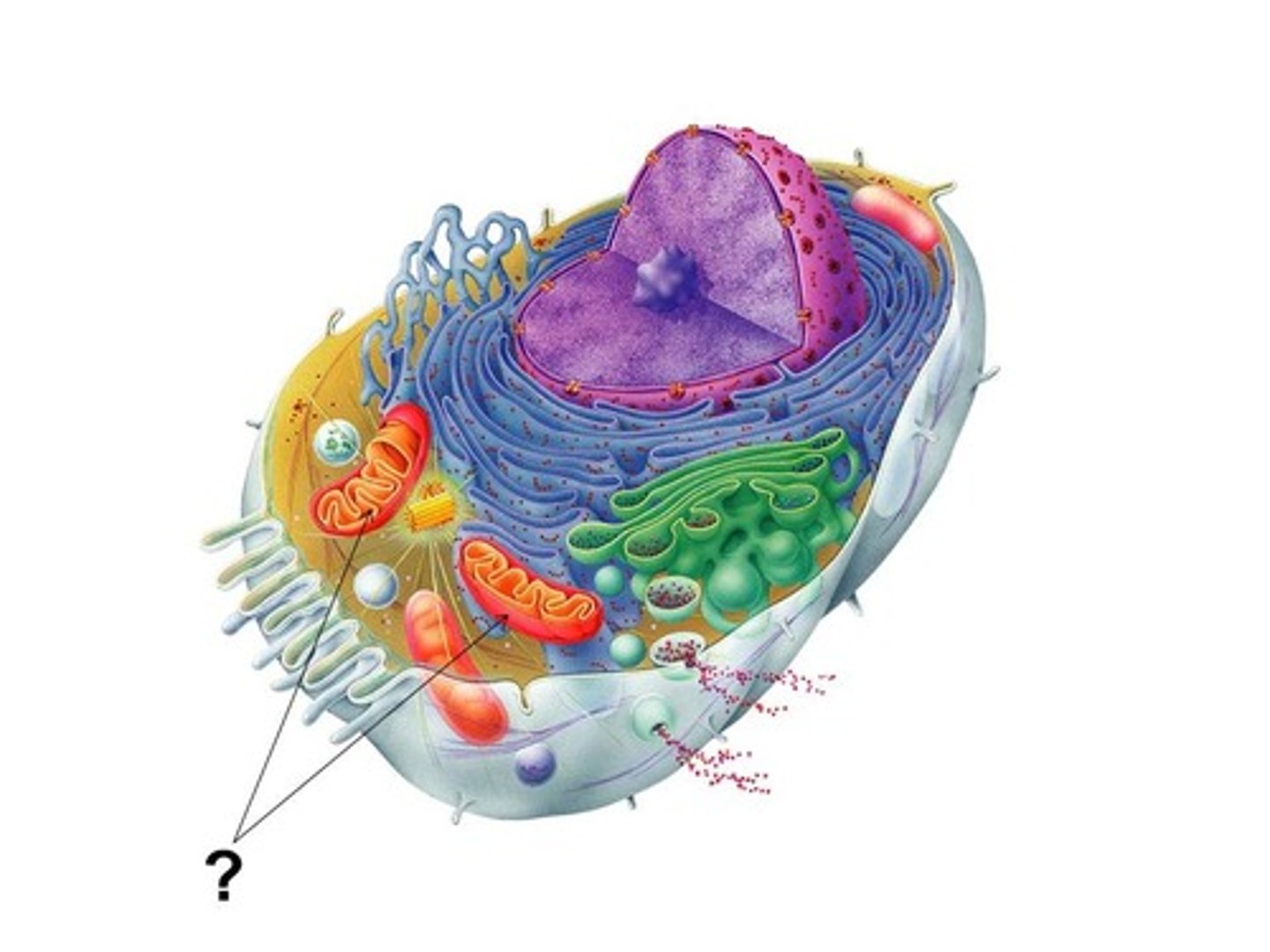
mitochondria
-organelles of plant and animal cells
-site of oxidative metabolism (generate ATP)
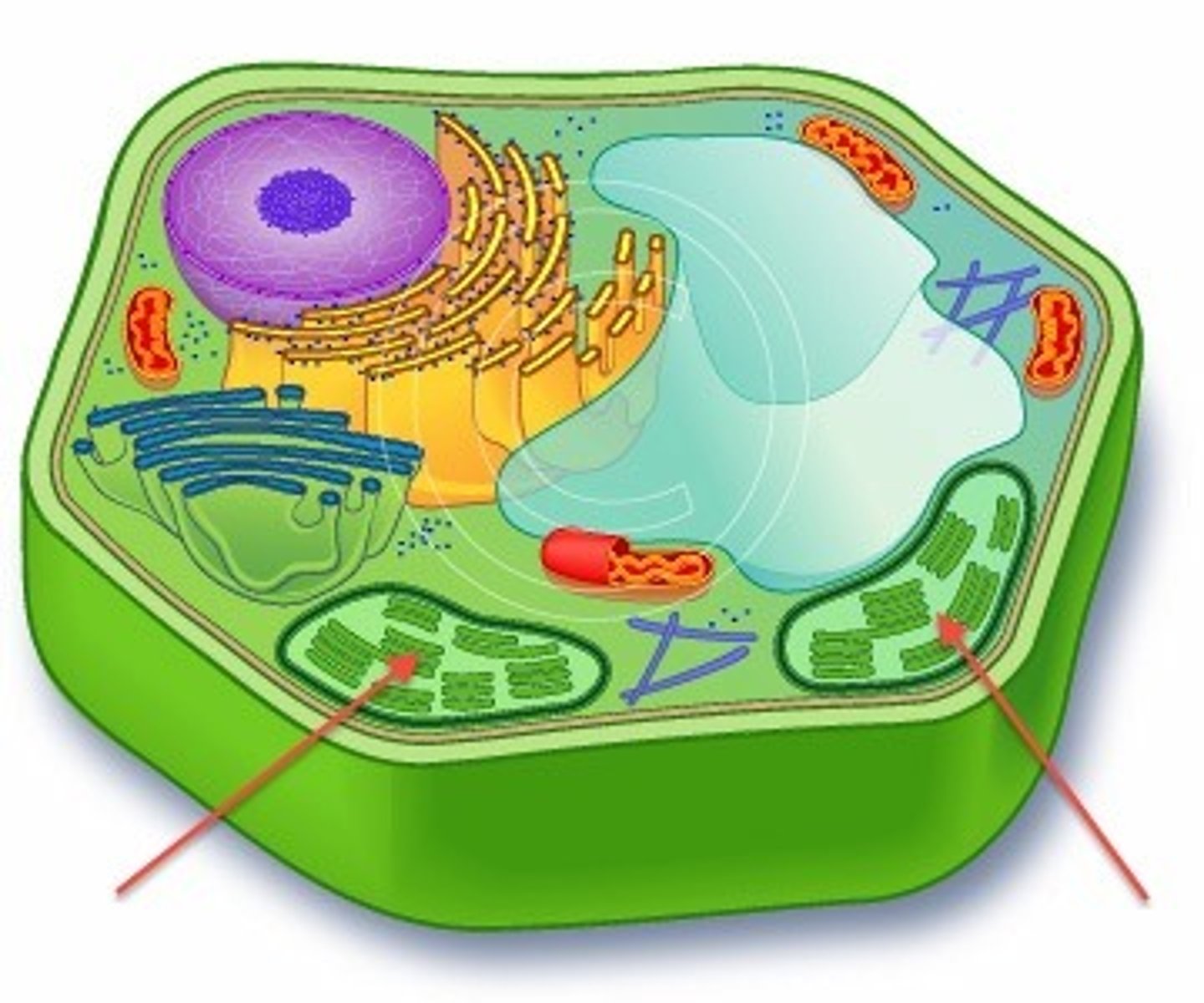
chloroplasts
-organelles of plant cells
-site of photosynthesis
lysosomes
-organelles of plant and animal cells
-site of digestion of macromolecules
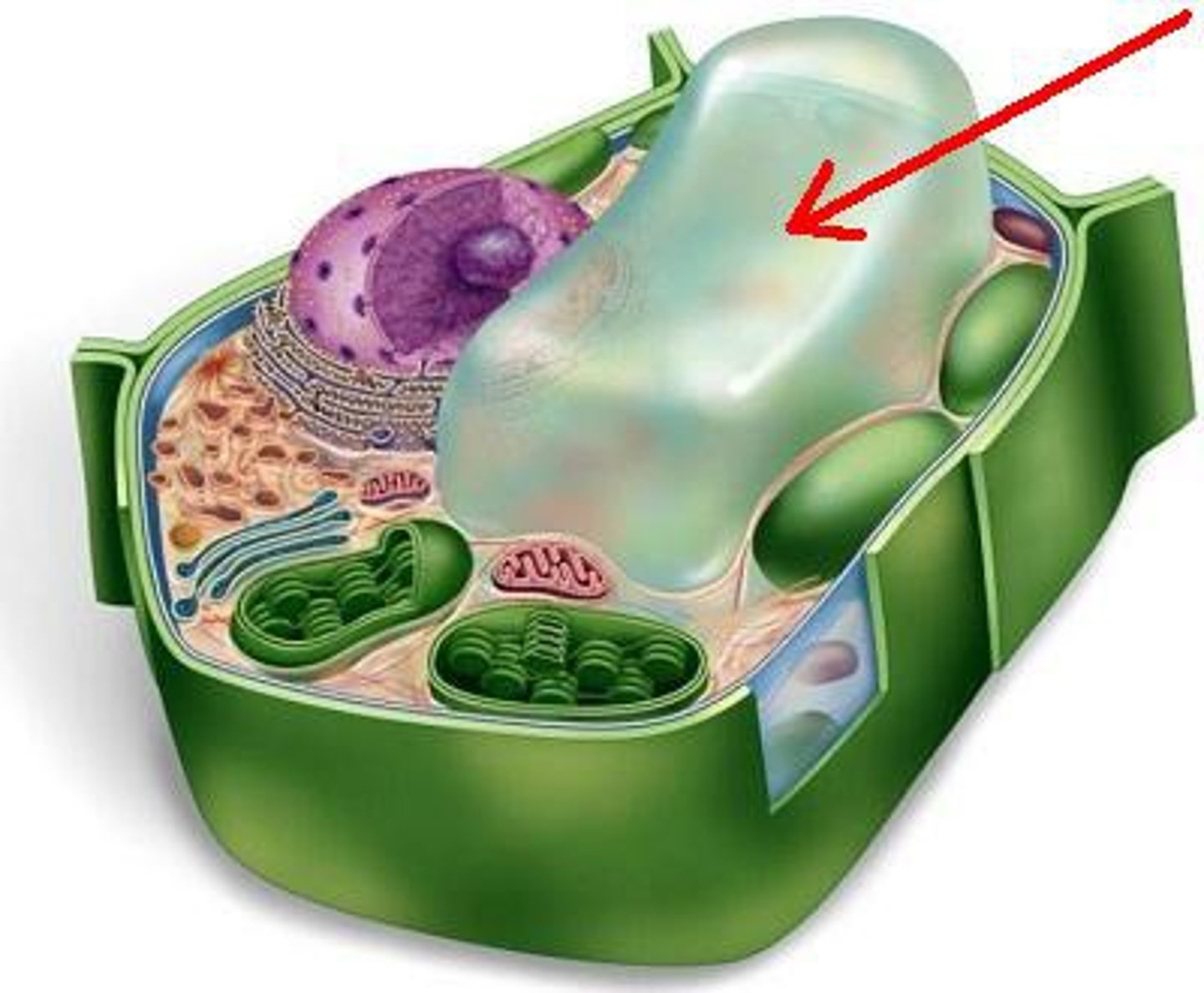
vacuoles
-organelles of plant cells
-site of digestion of macromolecules
-storage of waste products and nutrients
endoplasmic reticulum (ER)
-organelles of plant and animal cells
-network of intracellular membranes
-rough: processing and transport of proteins
-smooth: synthesis of lipids
golgi apparatus
-organelle of plant and animal cells
-site of further processing and sorting of lipids and proteins and sending to final destinations
-site of cell wall synthesis in plant cells
cytoskeleton
-organelle of plant and animal cells
-protein filaments extending through the cytoplasm
-provides structural framework and shape
-involved in movement of cell and organelles within the cel
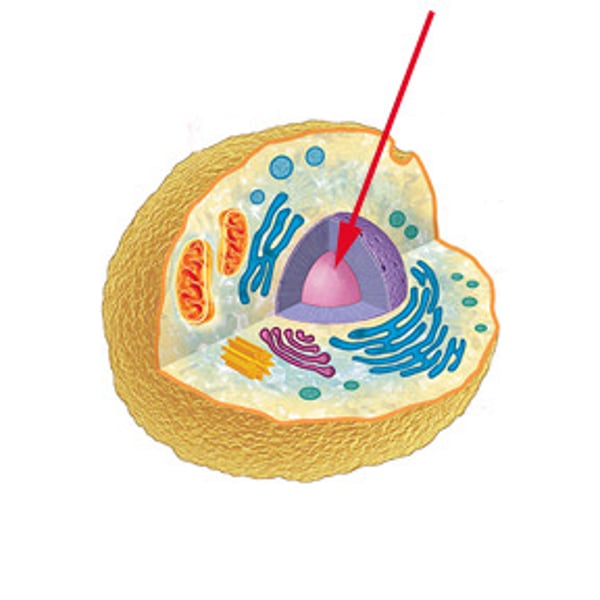
nucleus
-organelle of plant and animal cells
-largest organelle
-contains linear DNA
-site of DNA replication and RNA synthesis
DNA (deoxyribonucleic acid)
genetic material of all cells
-organized into genes which are the functional unit of inheritance that code for specific RNA and protein sequences
-composed of 2 polynucleotide chains that are joined by hydrogen bonds between complementary base pairs
protein
a polypeptide with a unique amino acid sequence
-created by the process of translation when RNAs that encode proteins use their nucleotide sequence to specify the order of amino acids
RNA (ribonucleic acid)
a polymer of ribonucleotides
-created by the process of transcription when the nucleotide sequence of DNA is copied
E. coli (escherichia coli)
-bacterial experimental model for studies of molecular genetics
-led to understanding of DNA replication, the genetic code, gene expression and protein synthesis
-simple and small genome (4000 genes)
-grows rapidly (divide every 20 mins)
-can create clonal populations (such as antibiotic resistant strains)
-require relatively few nutrients and can produce their own nutrients if necessary
yeast (saccharomyces cerevisiae)
-eukaryotic experimental model used to study eukaryotic cell biology (molecular biology)
-simplest eukaryotes (6000 genes) but still have typical characteristics of complex eukaryotes
-divide frequently (every 2 hours)
-can create colonies of mutants
-have been used to understand DNA replication, transcription, RNA processing, protein sorting and regulation of cell division in eukaryotes
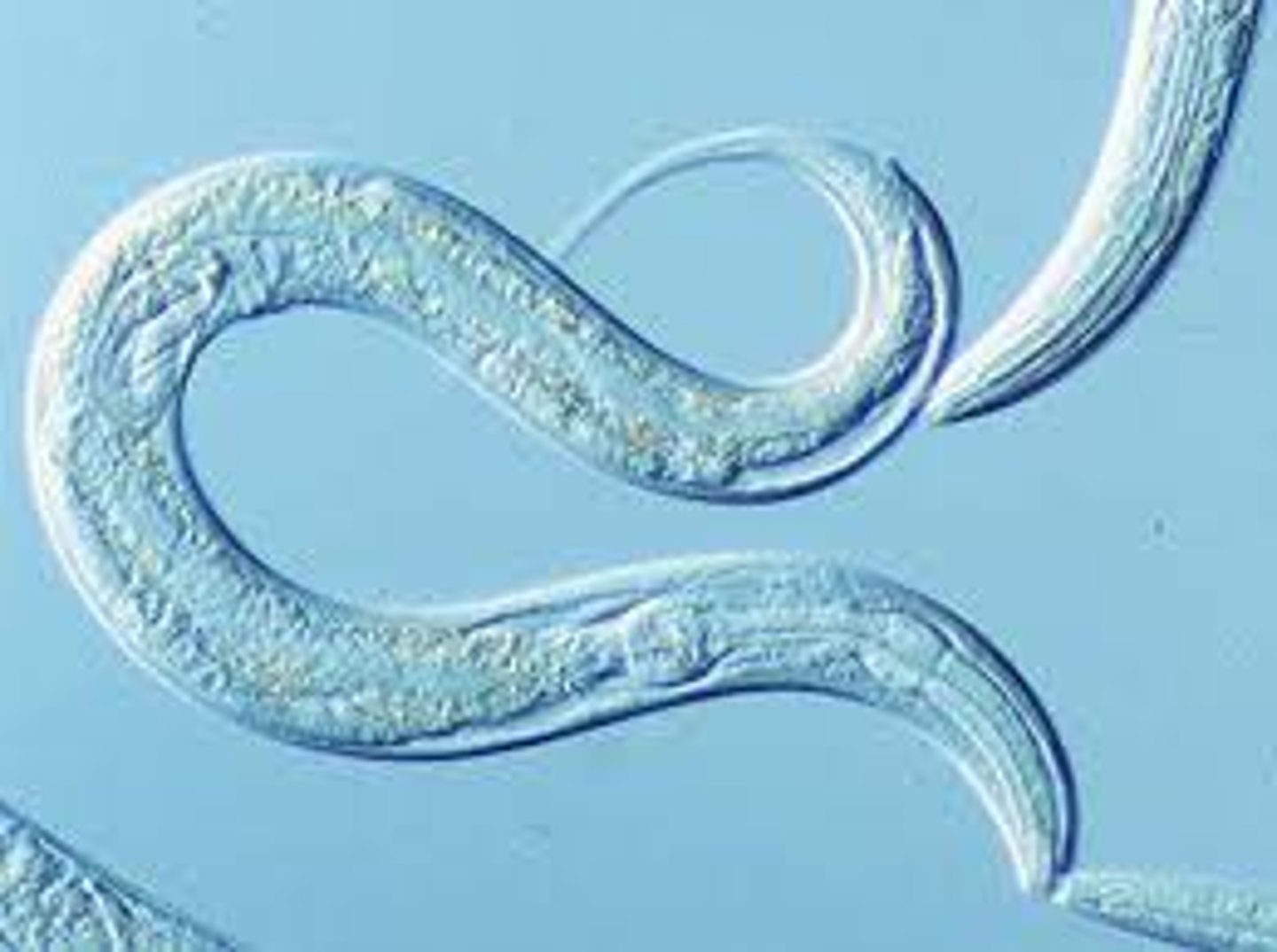
nematode (Caenorhabditis elegans)
-experimental model of multicellular organisms for animal development and cell differentiation
-relatively small and simple genome for multicellular organism, but same number of protein coding genes as humans
-easily grown and genetically manipulated in labs
-all cells in adult have been traced from embryonic lineage
-genetic studies have identified mutations for developmental abnormalities and characterization of genes for development and differentiation and applied to complex organisms
fruit fly (Drosophila melanogaster)
-model organism in developmental biology
-easily maintained and bred in labs with short reproductive cycle (2 weeks)
-useful for genetic experiments
-relationship between genes and chromosomes discovered in this species
-has led to discovery of genes that control development and differentiation and led to methods of molecular biology for gene analysis
-led to advances in molecular mechanisms that govern animal development that exist across all vertebrates
-mutations were observed for eye color and wing shape and breeding experiments explained segregation of genes on different chromosomes

mouse-ear cress (Arabidopsis thaliana)
-model organism for plant molecular biology and development
-small genome with representative characteristics of most plants
-easy to grow in lab and able to be genetically manipulated
-led to identification of genes involved in plant development
-shows similarities and differences in development of plants and animals

zebrafish
-model organism for vertebrate development
-easy to maintain in labs and reproduce rapidly (3-4 months)
-embryos are easy to study bc they're transparent and develop outside of mother
-used to identify developmental mutations and bridge gap to humans (vertebrates)

mouse
-model organism for genetic analysis of mammals applied to humans
-technical difficulties of studying their genetics
-have recently developed ability to engineer the genome to include mutant genes to study the function of the genes
-mutations in homologous genes to human result in similar developmental defects to humans

embryonic stem cells (ES cells)
-approach to studying cells of multicellular organisms is growing isolated cells in culture
-allowed studies of mammalian cell biology
-led to understanding of mechanisms of DNA replication, gene expression, protein synthesis and processing and cell division
-allowed study of signaling mechanisms that control growth and differentiation within organisms
-wide variety of animal cells can be grown
frog (Xenopus laevis)
-model organism for studies of early vertebrate development
-produce large number of eggs that are large in size
-make lab study and biochemical studies easy

light microscopy
-basic tool of cell biologists is microscope that magnifies objects about 1000x their size
-allows visualization of cells and some of their organelles
-tool that led to discovery of cells by robert hooke
-antony van leeuwenhoek used to view variety of cells (RBCs, sperm, bacteria)
-led to development cell theory that cells can only come from existing cells
bright-field microscopy
-tool used to visualize cells in which light passes directly through the cell
-cells are preserved with fixatives and stained (CANNOT be used to study living cells)
phase-contrast microscopy and differential interference-contrast microscopy
-tool that converts variation in density / thickness to differences in contrast
-useful for seeing living cells
-videos can be used to visualize the movement of organelles
video cameras and computers
-tools used for image analysis and processing
-enhance the contrast of images
fluorescence microscopy
-tool used for molecular analysis
-fluorescent dye is attached to dead or living cell and can either absorb or emit light
green fluorescent protein (GFP)
-tool used to study living cells by fusing it to a protein of interest using recombinant DNA
-tagged protein is expressed in cells and detected by fluorescence microscopy
fluorescence resonance energy transfer (FRET)
-tool used to visualize cells in which 2 proteins are coupled to different fluorescent dyes and light emitted by one excites the second
-used to study interactions between proteins in cells
confocal microscopy
-tool using a single point in the specimens to analyze fluorescence and increase contrast and detail
-produces 2D images of plane of focus with very sharp images
multiphoton microscopy
-localized excitation of a fluorescent dye at a point where a laser beam is focused
-produces 3D images of living cells with minimal damage to specimen
electron microscopy
-achieves greater resolution than light microscopy with short wavelength of electrons
transmission electron microscopy
-specimens are fixed and stained with slats of heavy metals to provide contrast by scattering electrons
-beams of electrons is passed through the specimen
-used to determine specific locations of proteins within cells
scanning electron microscopy
-surface of cells is coated with heavy metals and beam of electrons is used to scan across the specimen (NOT through it)
-produces a 3D image of cells
-can use positive or negative stain
freeze fracturing
-specimens are frozen with liquid nitrogen and fractured with knife blade to split lipid bilayer
-allows interior faces of cell membrane to be visualized
subcellular fractionation
-organelles are isolated from the cell to determine the function
-usually accomplished by differential centrifugation to separate the components of the cell and then they are studied with biochemical treatments
differential centrifugation
cell components are separated by size and density
-ultracentrifuge spins at high speeds to accomplish this
water
-most abundant molecule in all cells
-causes cell membranes of phospholipids to assemble
organic molecules
-carbohydrates
-lipids
-nucleic acids
-proteins
-all present in all cells
carbohydrates
-simple sugars and polysaccharides
-serve as major nutrients of cells and surface markers
-composed of C, H and O
lipids
-used for storage of energy, cell membranes and cell signaling
-fatty acids are stored as triaglycerols or triglycerides
-phospholipids primarily make up cell membranes, also include glycolipids and cholesterol
-steroid hormones (estrogens and testosterone) are derivatives of cholesterol
nucleic acids
-principal informational molecules in cells
-DNA is composed of purines A and G and pyrimidines C and T
-RNA is composed of A and G and C and U
proteins
-serve many functions in cells such as structure, transport and storage, transmission, defense and enzymatic activity
-are all composed of 20 different amino acids
-joined by peptide bonds
-4 levels of structure
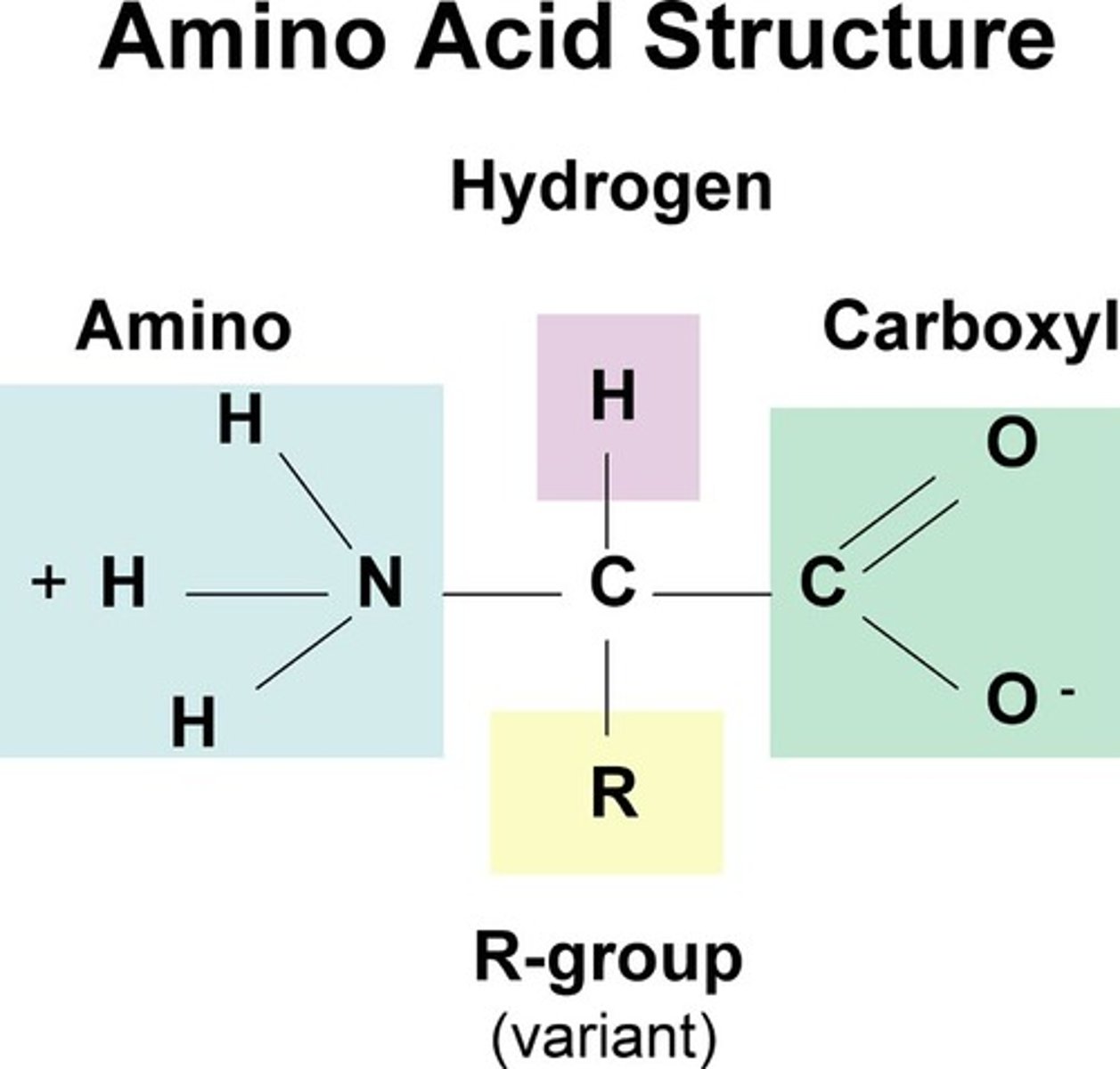
amino acids structure
carbon atom bonded to carbonyl group (COO), amino group (NH3), a hydrogen atom and a side chain
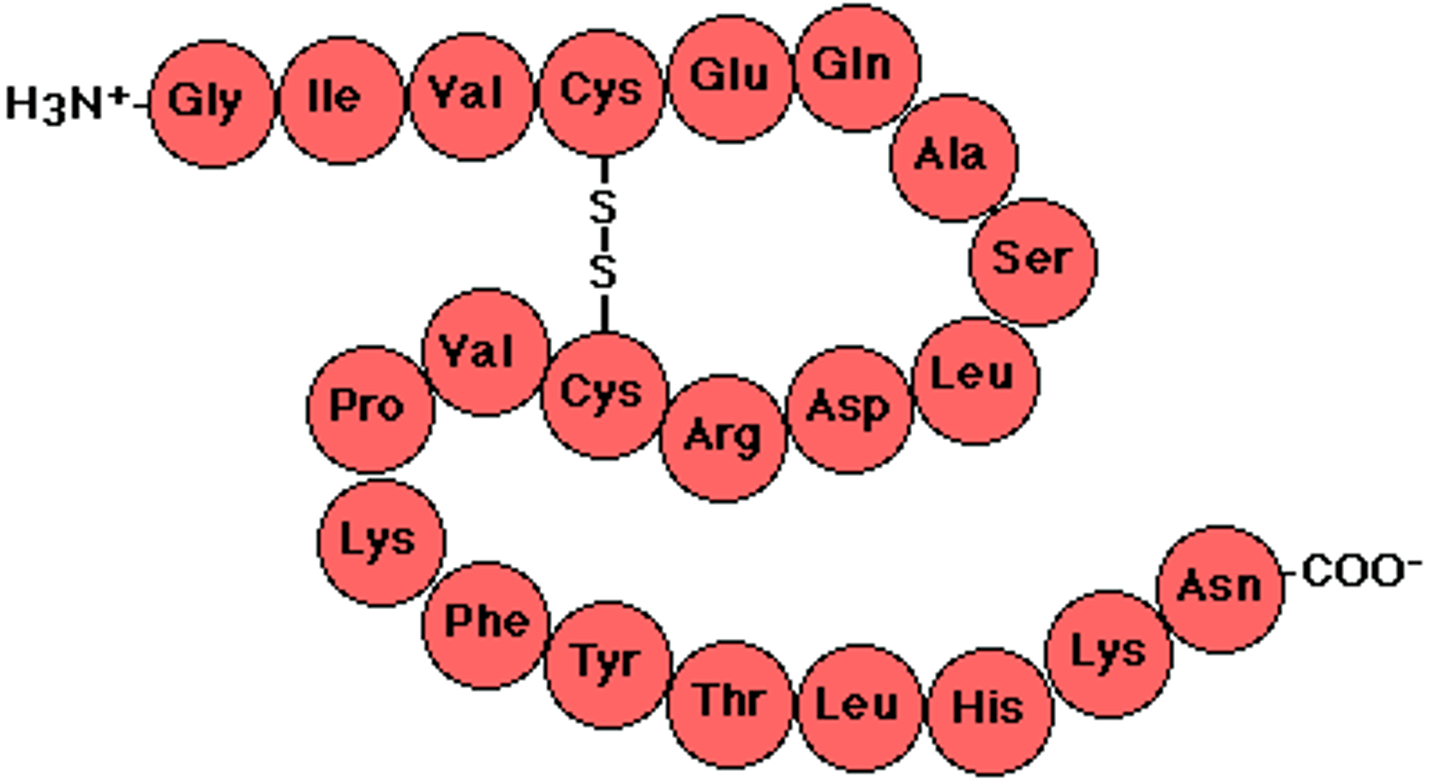
polypeptide structure
linear chains of amino acids joined by peptide bonds, usually 100's to 1000's of amino acids long
-considered the primary structure of proteins
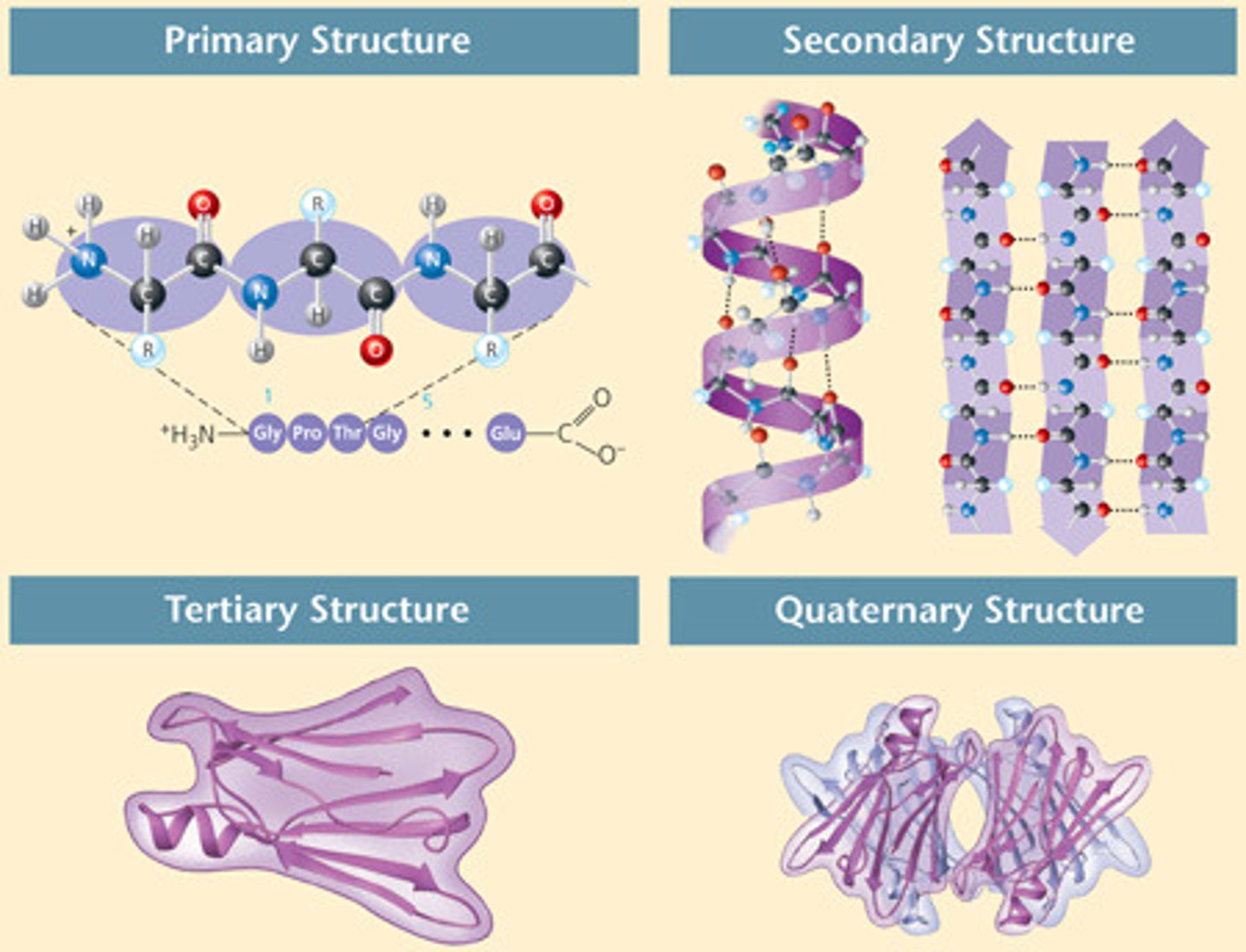
protein structure
-primary: polypeptide chain
-secondary: arrangement of polypeptide into alpha helix or beta pleated sheet
-tertiary: overall folding of a single polypeptide due to interactions of side chains (domains)
-quaternary: interactions between different polypeptide chains in proteins that are composed of more than one chain
enzymes
catalytic proteins that increase the rate of chemical reactions without being consumed or altered and without altering the chemical equilibrium of products and reactants
-may require coenzymes
mRNA (messenger RNA)
carries information from DNA to the ribosomes
nucleotides
-units that become linked by phosphodiester bonds to become RNA and DNA
-consist of purine and pyrimidine bases linked to phosphorylated sugars (2'-deoxyribose or ribose)
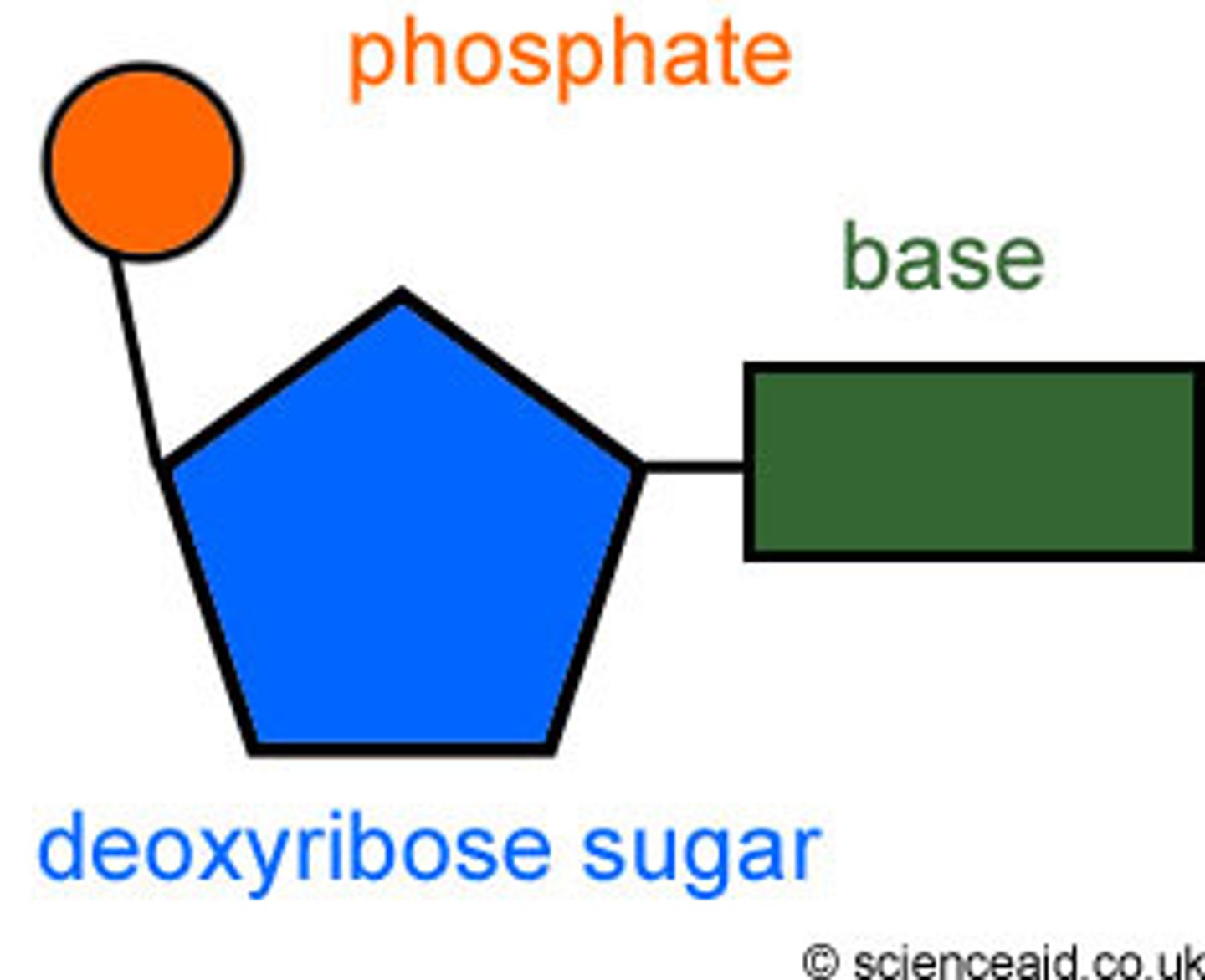
DNA
-genetic material in cells
-in eukaryotes is contained in the nucleus
-double stranded helix
tRNA (transfer RNA)
-RNA molecule that functions as adaptor between amino acids and mRNA during protein synthesis
rRNA (ribosomal RNA)
-the RNA that is part of the structure of ribosomes
-involved in protein synthesis
monosaccharides
serve as main source of energy for cells (specifically glucose)
-single sugar
-usually present in ring form
disaccharides
2 sugars linked together by glycosidic linkage
oligosaccharides
polymer of only a few sugars
-frequently linked to proteins and play role in protein folding
-may function as cell surface markers
polysaccharides
polymer of hundreds or thousands of monosaccharides
-function in storage of energy and markers in cell recognition
-glycogen and starch are storage forms of carbohydrates used for energy in animal and plant cells
-cellulose in plants and chitin in certain animals are structural carbohydrates
origin of cells
-emerged 3.8 billion years ago
-likely developed from organic molecules
-surrounded by bilayer that formed spontaneously in water
-all modern cells descended from single common ancestor
cellular mechanisms of generating energy
-glycolysis: produces small amount of ATP in anaerobic environment
-photosynthesis: harnesses energy from sunlight to produce own organic molecules and produces oxygen in atmosphere
-oxidative metabolism: yields 36-38 ATP
resolution
ability to distinguish objects separated by small distances on microscope
super resolution light microscopy
microscopy technique that increases the resolution of fluorescence microscopy to 10-100 nm
density gradient centrifugation
organelles are separated by sedimentation through a gradient of a dense substance
velocity centrifugation
particles of different sizes sediment through the gradient at different rates
equilibrium centrifugation
separates subcellular components on basis of buoyant density
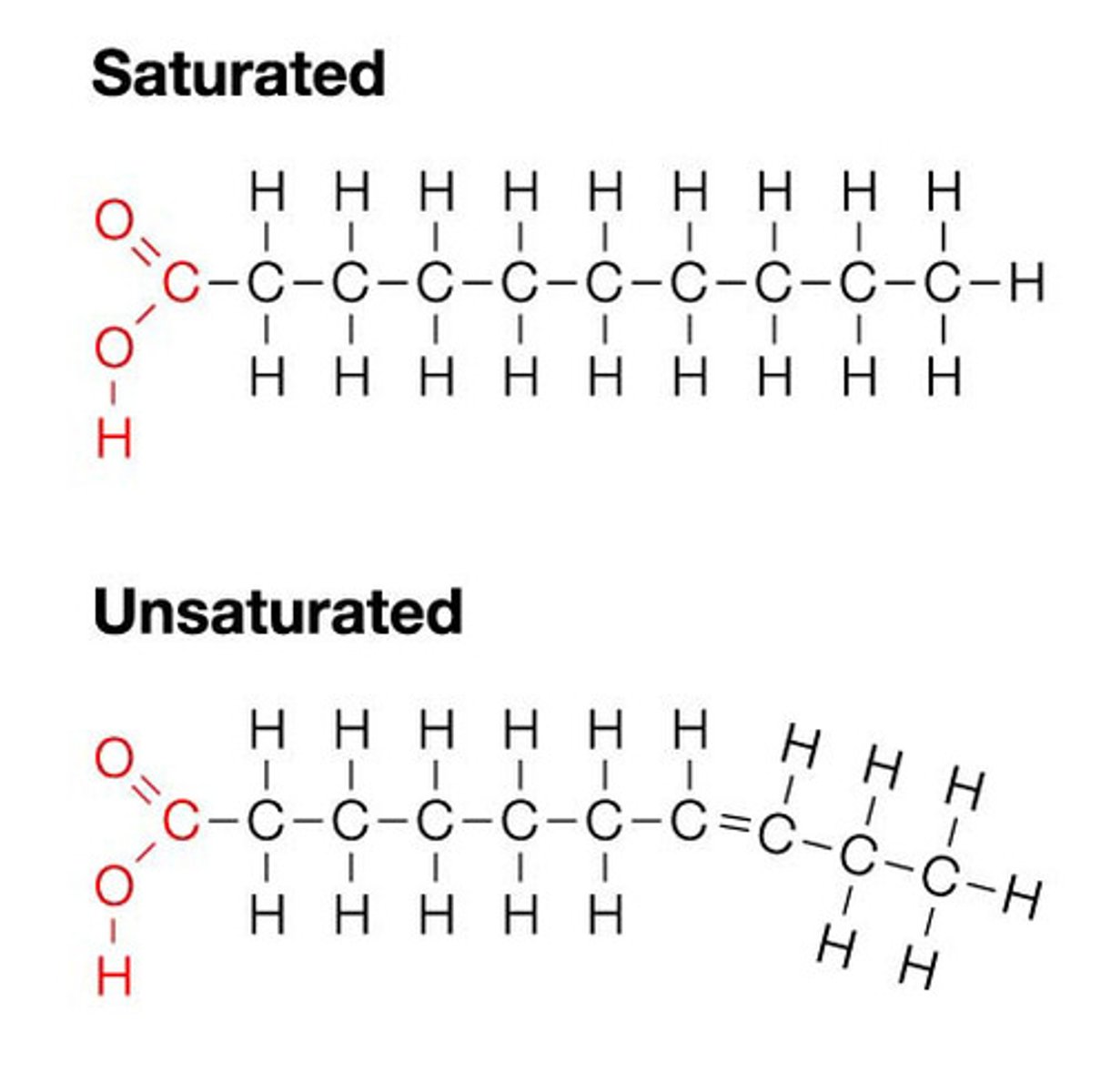
fatty acids
-simplest lipids consisting of long hydrocarbon chain with carboxyl group (COO) at one end
-may be saturated (single bonds between C's) or unsaturated (double bonds between some C's)
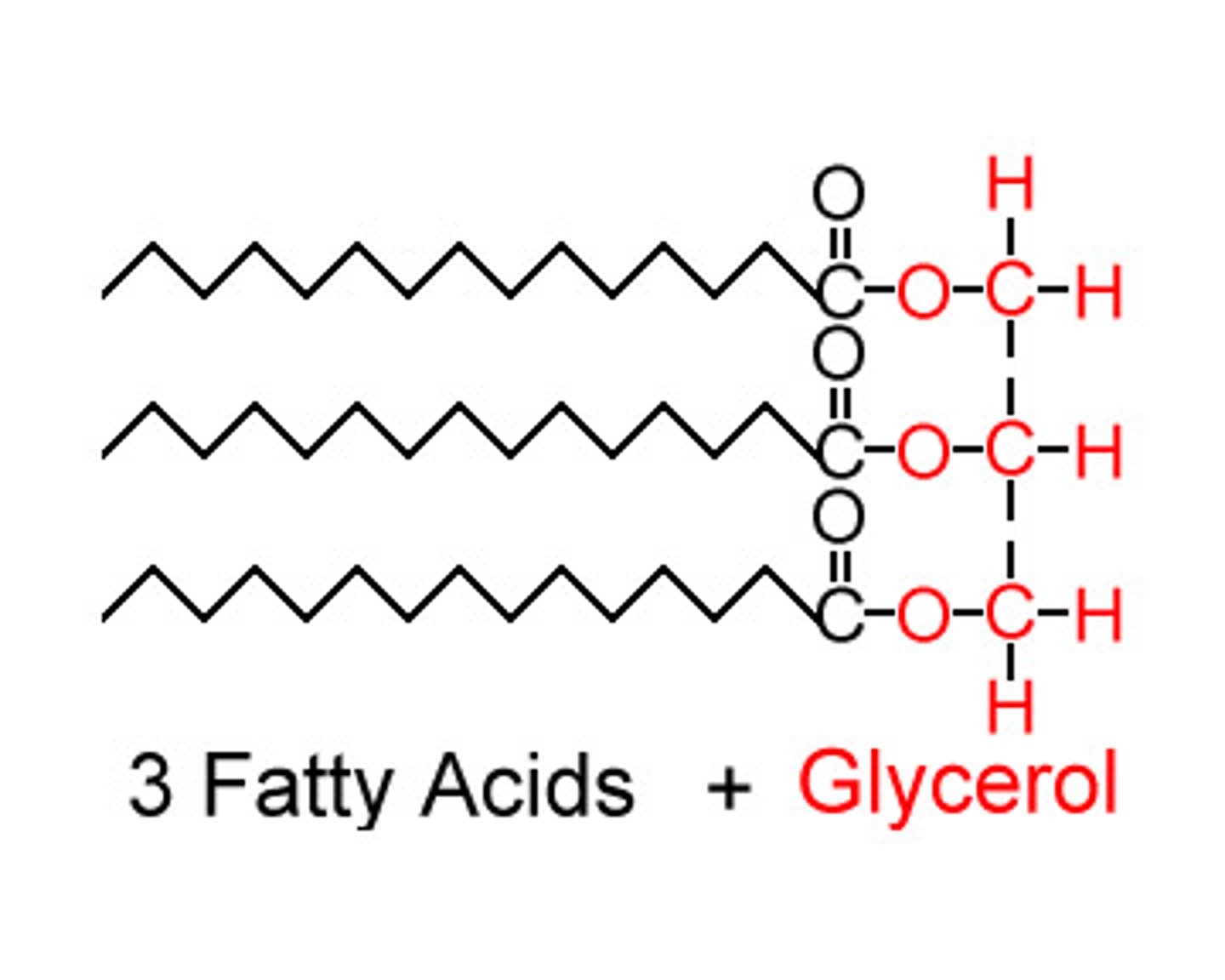
triaglycerols (triglycerides)
-storage form of fatty acids
-three fatty acids linked to a glycerol molecule
-can be broken down for energy when needed
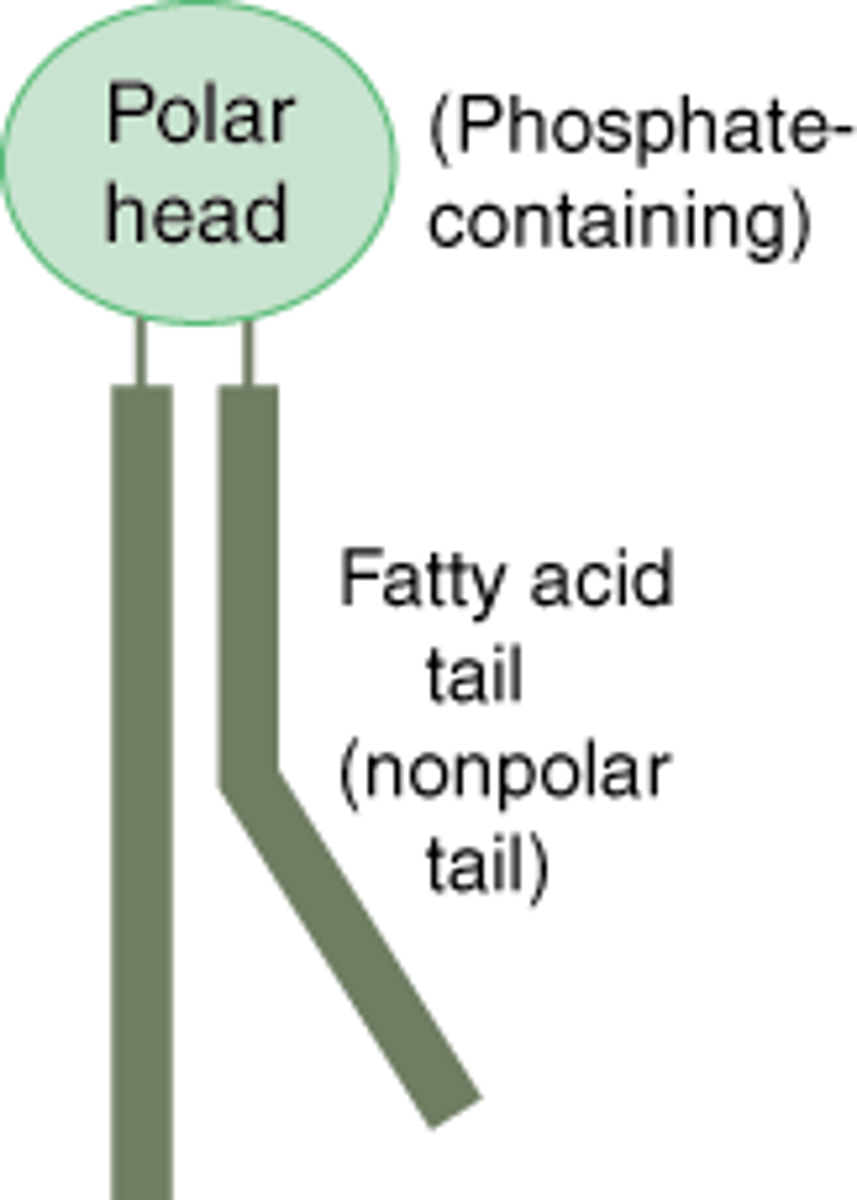
phospholipids
-amphipathic molecules that aggregate to form lipid bilyaers
-two fatty acids joined to a polar head group
-polar head group may be glycerol bound to phosphate group or other small polar molecule
-sphingomyelin is only non-glycerol head group, is attached to a serine
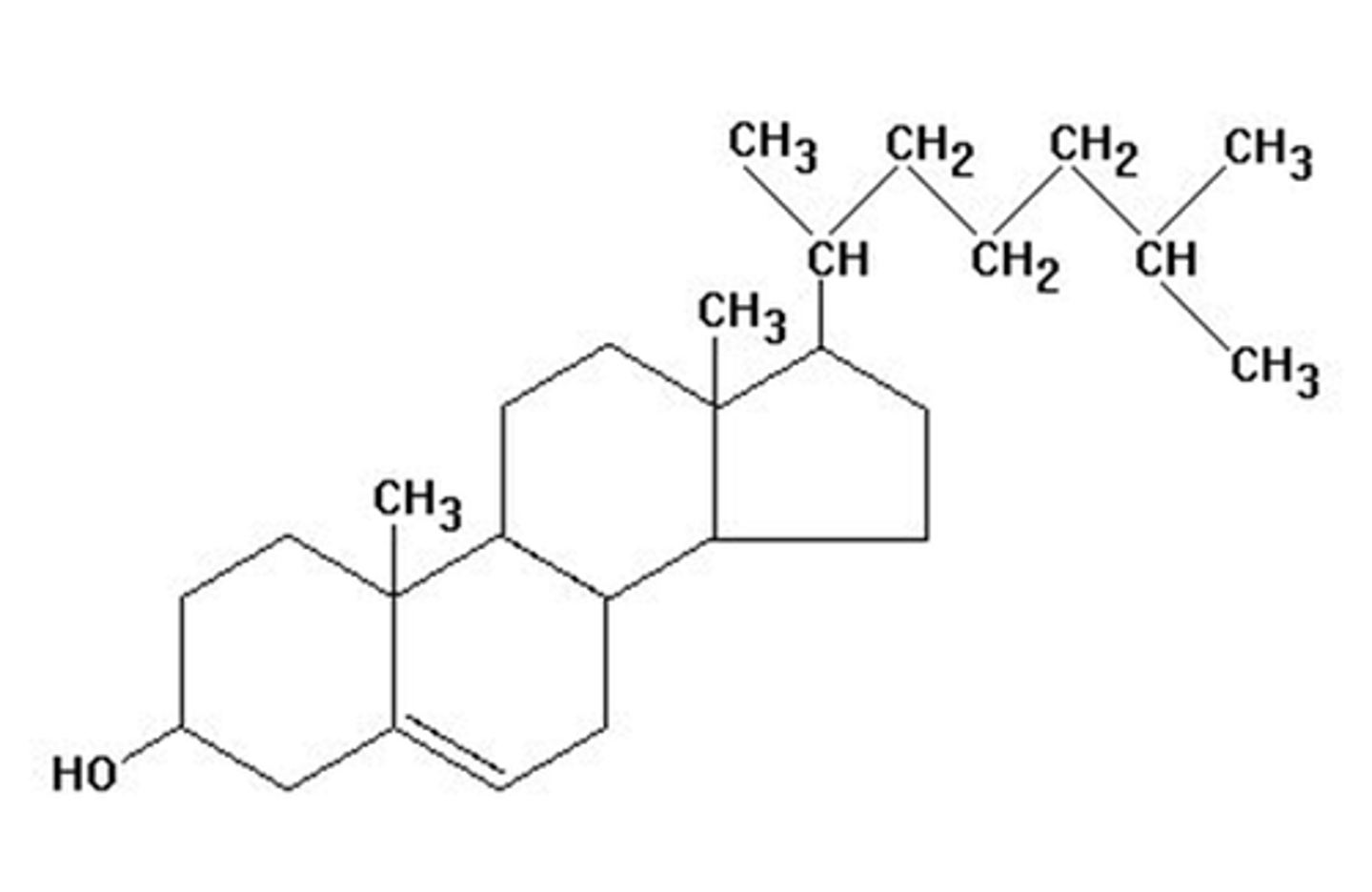
cholesterol
-four hydrocarbon rings with one hydroxyl (OH) group that is weakly polar making the molecule amphipathic
-present in cell membranes to maintain fluidity
-steroid hormones are derived from this
glycosidic bonds
bonds that are formed between monosaccharides during dehydration reactions
phosphodiester bonds
bonds that are formed between nucleotides
-always synthesized in the 5' to 3' direction during polymerization
peptide bonds
bonds that form between amino acids to create polypeptides
cell membrane
-composed of phospholipids, with insertions of proteins, glycolipids and cholesterol
-present as plasma membrane and membranes surrounding organelles such as nuclei, mitochondrion, etc.
-selectively permeable
-molecules in the membrane freely rotate and move
gene
-pair of inherited factors (i.e. YY) that are carried on chromosomes
-functional unit of inheritance that corresponds to a segment of DNA that encodes a polypeptide or RNA molecule
allele
-single gene copy (i.e. Y or y)
-one is inherited from each parent to produce a full gene
genotype
-the genetic composition of an organism that determines the physical appearance
-i.e. it would either be Yy or YY or yy
phenotype
-the physical appearance resulting from the combinations of gene alleles
-i.e. the combination Yy would cause the organism to appear yellow
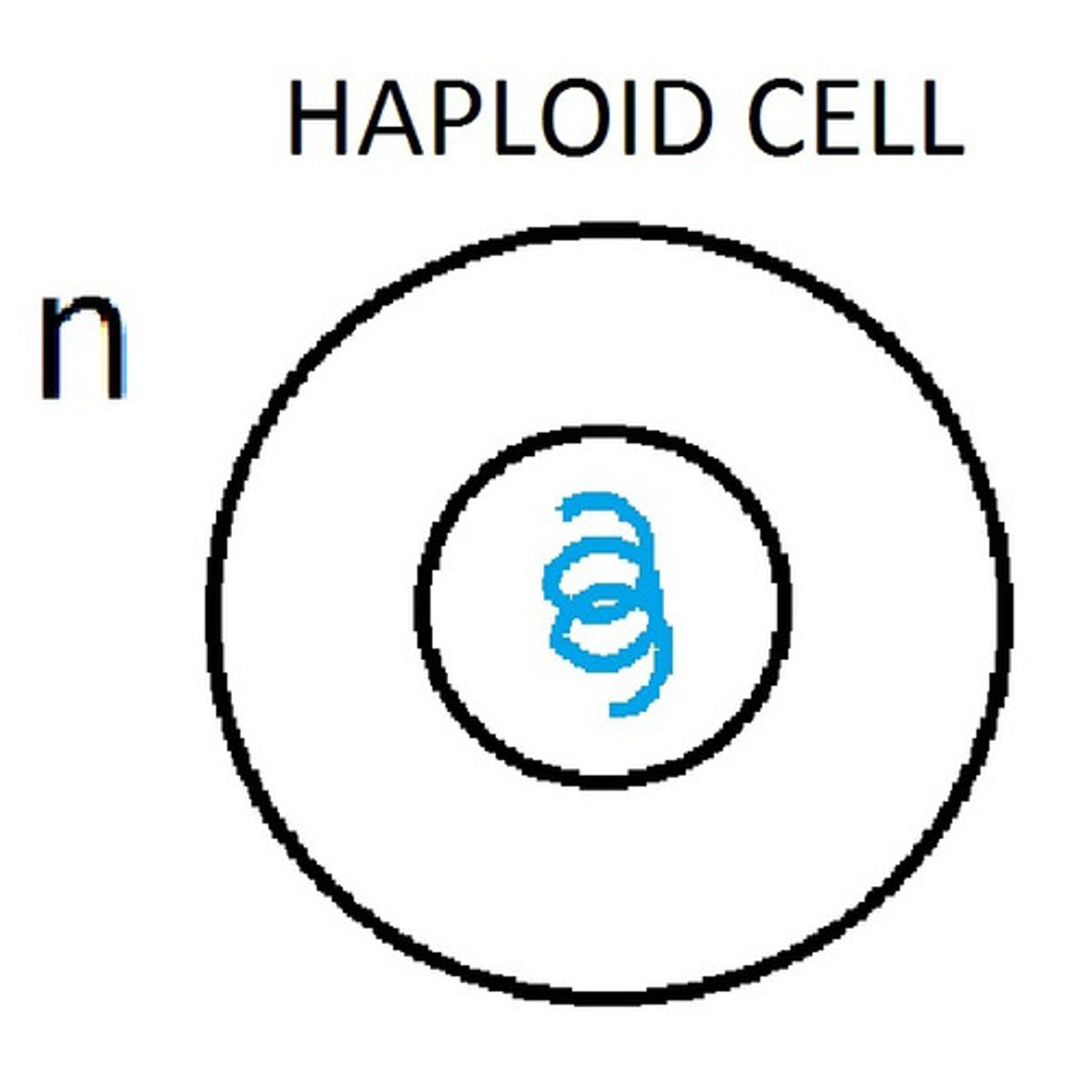
haploid cells
-a cell that has one copy of each chromosome
-sex cells (eggs and sperm)
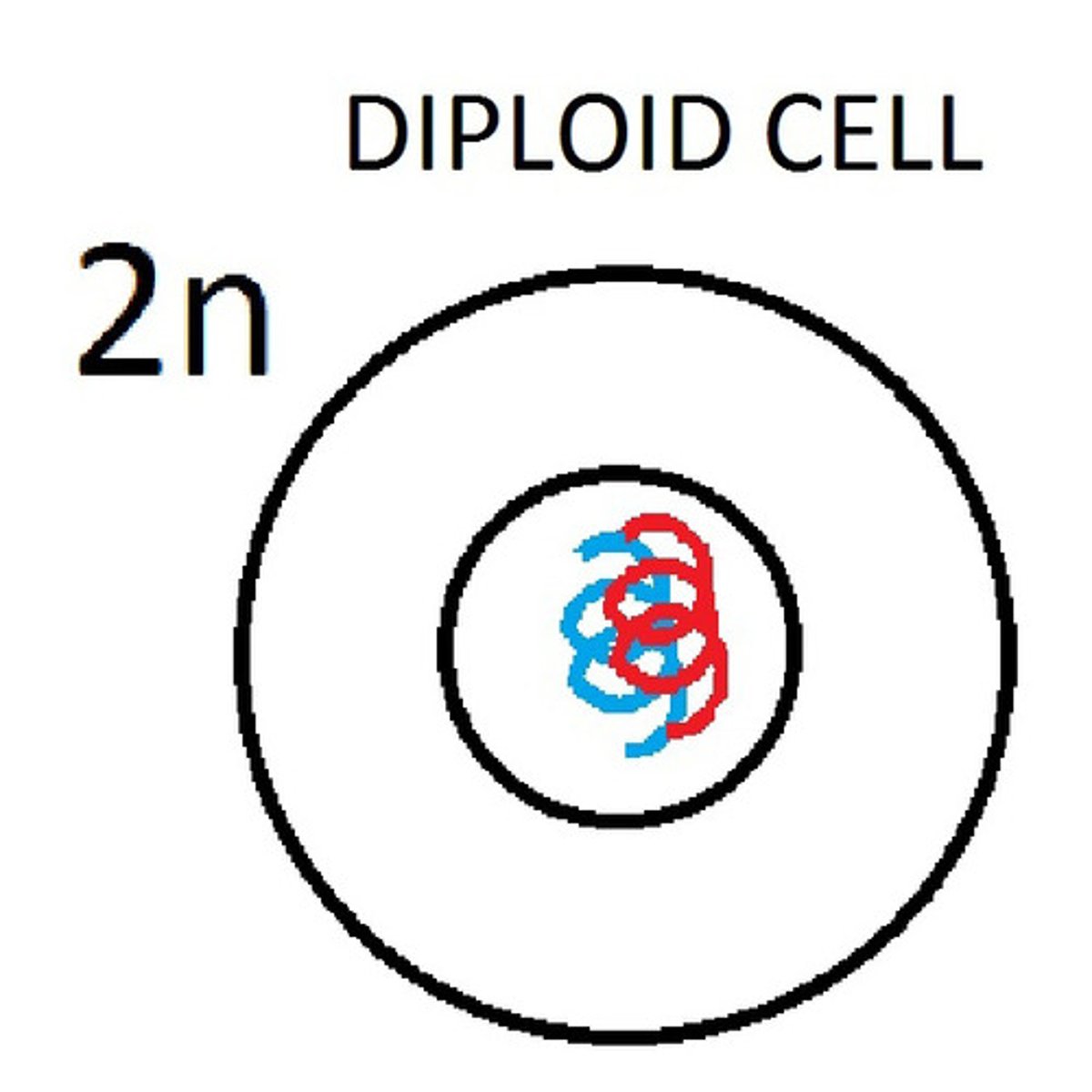
diploid cells
-a cell that carries two copies of each chromosome, one from each parent
-all body cells (except sex cells)
gregor mendel
-began the study of genetics
-breeding experiments with pea plants looking at color of seeds to observe patterns of inheritance
-determined that yellow was the dominant trait (YY) and green was recessive (yy), so even hybrids (Yy or yY) appear yellow
relationship between genes and proteins
-phenylketonuria results from a genetic defect in region that codes for the enzyme (protein) needed to metabolize the amino acid phenylalanine
-revealed that specific gene regions of DNA code for proteins that function in the body, and when there is error in the DNA there is errors in the proteins it creates
semiconservative replication
when DNA is replicated the double strands are opened and each individual strand serves as template for new strand being formed
-new DNA will have 1 new strand and one strand of parent DNA
genetic code
-discovered by Nirenberg and Khorana
-nucleotide "triplet" codons (3 nucleotides in a row) code for specific amino acid
-there are many codons that can code for a single amino acid
-DNA sequence determines RNA sequence which codes for amino acids
-there are 64 total codons, 3 are for STOP and 61 are for amino acids
reverse transcription
-enzyme catalyzed synthesis of DNA formed from an RNA template
-occurs in cells and is critical for replicating ends of eukaryotic chromosomes
-responsible for transcription from DNA of one chromosome to another
-reverse transcriptase generates DNA copies from RNA
recombinant DNA
-produced by PCR technique of inserting a DNA segment of interest into a DNA vector that is capable of independent replication in a host cell
-allows scientists to isolate, sequence and manipulate individual genes derived from any kind of cell
-restriction endonucleases are used to cleave DNA at specific sequences and insert DNA of interest
restriction endonucleases
enzymes that cleave DNA at specific sequences, used in PCR
-discovered in bacteria as a defense against foreign DNA
gel electrophoresis
-segments of nucleic acid gain a negative charge and are positioned in a gel between 2 electrodes
-segments of nucleic acid migrate through the gel, with the smaller molecules moving most rapidly toward the positive electrode
-allows DNA molecules that were cleaved by restriction endonuclease to be isolated
cosmid vectors
-PCR vector used with larger DNA fragments
-contains bacteriophage lambda sequences, origins of replication and genes for antibiotic resistance
-allows DNA to be replicated as plasmids within bacterial E. coli cells
bacteriophage P1 vectors
-PCR vector used with larger DNA fragments
-recombinant molecules are packaged in vitro into the p1 bacteriophage and is replicated as plasmids inside of E. Coli
P1 artificial chromosome (PAC) vectors
-PCR vector used with larger DNA fragments
-recombinant molecules have the P1 sequence but are replicated directly as plasmids inside of E. Coli
-NOT introduced into bacteriophage
bacterial artificial chromosome (BAC) vectors
-PCR vector used with larger DNA fragments
-DNA is introduced into a naturally occurring plasmid of E. coli, the F factor
yeast artificial chromosome (YAC) vectors
-PCR vector used with larger DNA fragments
-DNA of interest is grown in yeast origin of replication allowing creation of linear chromosomes
polymerase chain reaction (PCR)
-DNA polymerase is used in vitro for exponential replication of a DNA segment of interest using a heat stable enzyme
-DNA sequence is amplified through use of a known DNA sequence to begin replication at the beginning of the desired sequence
real time PCR (RT-PCR)
-PCR used to quantitatively determine the amounts of specific DNA or RNA molecules in an inital sample
-fluorescent dye attaches to double stranded DNA is incorporated into PCR reaction and fluoresces when double stranded DNA is produced
-fluorescence is measured after each replication to measure the amount of DNA produced each cycle, which tells how much was initially present
nucleic acid hybridization
-double stranded DNA is denatured at high temperatures causing strands to separate
-strands are incubated and allowed to re-bind according to complementary base pairing
-used to detect DNA or RNA sequences that are complementary
southern blotting
-used to detect specific genes in DNA
-DNA is digested with restriction endonuclease and separated by gel electrophoresis
-gel is blotted with thin membrane or filter to create replica of gel
-filter is incubated with a labeled probe that hybridizes to DNA fragments that contain the complementary sequence to the fragments
northern blotting
-used to detect specific genes in RNA (studies of gene expression)
-RNAs are separated by gel electrophoresis
-gel is blotted onto a filter and filter is incubated with a labeled probe that hybridizes to complementary RNA fragments
pneumonococcus
-showed evidence that DNA is the genetic material
-activity of the transforming principle was steopped when enzymes were digested, but could be regained due to creation of more proteins from DNA
-when DNA was digested all activity stopped because no new enzymes could be created
watson and crick
used xray crystallography to determine double helix with sugar phosphate back bone and hydrogen bonding formation of DNA