BIOL215 Midterm
1/98
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
99 Terms
Evidence for Evolution
Uniformity of life, common descent, fossils, and extinction
Adaptation
Fit between organism and its environment
Lack of fit → selection can improve fit
Technical Definition: heritable phenotype that increases fitness and is caused by natural selection in the current environment
Natural Selection
One of four mechanisms for evolution
One lineage replicates faster than others
Phylogeny
Evolutionary history of a lineage or lineages (populations, genes, or species)
Phylogenetic Tree
visual representation of a phylogeny
Monophyletic Group
group of organisms that includes a single common ancestor and all of its descendants, forming an unbroken line of evolutionary descent
Clade
monophyletic group within a phylogenetic tree
includes all species descended from a common ancestor
Paraphyletic Group
leaves out some taxa sharing a common ancestor
Polyphyletic Group
includes taxa descended from multiple common ancestors
Characters
Identifiable heritable traits
Character States
Condition of the character
Ex: Character—wings
Character States— present, absent
Described as ancestral and derived
Outgroup
Species outside of the clade
Shares a common ancestor with monophyletic clade on interest
Help identify synapomorphies
Synapomorphy
Shared derived character state, homologous characters (inherited from common ancestor)
Homoplasy
Character state similarity not due to common descent
Convergent evolution: independent evolution of similar trait
Evolutionary Reversals
reversion back to an ancestral character state
ex: swimming in whales
Maximum Parsimony
fewest evolutionary steps is preferred
more than one tree is equally parsimonious → consensus tree
logistically most likely to happen
Polytomy
relationships between species are uncertain
Maximum likelihood
Statistical approach to deciding on a tree
Range of trees is generated → most likely tee selected based on a particular model of evolution
Purifying Selection
selection wants to preserve function → evolve slowly
removes deleterious mutations and genetic variants from a population
ex: exons
Maximum Parsimony in Genetics
accounts for variable rates of evolution and homoplasy → more weight to slowly changing regions and less to rapid changing regions '
depends on interest in deep time or recent time
Deep Time
slowly evolving genes
Recent time
rapidly evolving genes
Bootstrapping
Used when have multiple equally parsimonious trees
Resample and reconstruct trees → resampled tres are similar is strong support for observed tree
Value 0-1 (1 is most likely)
Distance-Matrix Methods
more genetically similar → more likely to be closely related
branch lengths indicate number of changes
Models of Molecular Evolution (simple & complex)
Simple: all sites equally likely to mutate
Complex: different rates for nonsynonymous v synonymous, transitions v transversions, → genes evolve at different rates
Bayesian Method
Start with model and tree, slightly change tree many times → creates probability distribution of possible trees
Phylogenetically Independent Constrasts
Series of comparisons between nodes and tips of a phylogeny
Synonymous mutations
Don’t alter amino acid sequence, selectively neutral
Nonsynonymous mutations
Change amino acid sequence → subject to selection
Neutral Theory of Molecular Evolution
most evolution at molecular level is neutral and caused by random processes associated with genetic drift → neutral mutations become fixed in lineages at a regular rate
Selectionist Theory
Mutations are either advantageous or deleterious, selection is what drives molecular evolution/genetic diversity
Neutralist Theory
Most mutations are neutral, and genetic drift is the main cause of evolution
Selective Sweep
a beneficial allele will fix far more quickly than a neutral allele since its selected for → dip in genetic diversity at chromosome position where it becomes fixed
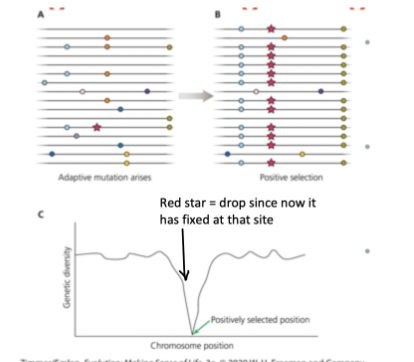
Genetic Hitchhiking
Mutations linked physically (right next to) selected genes also have very high frequencies
Eventually broken up by recombination in sexual reproduction over time
Dispersal
Movement of populations from one region to another
Limited or no return
Vicariance
Formation of geographic barriers that divide a once-continuous population
Anagenesis versus Cladogenesis
Ana: Creation of new species through gradual, step-wise change
Clado: Faster, lineage splitting
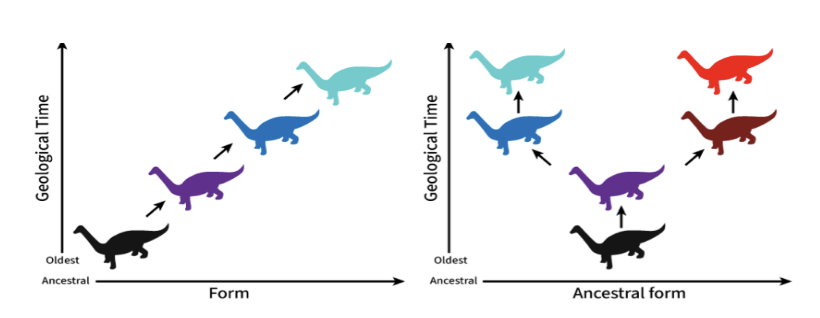
Gradualism
slow, gradual morphological changes over time → speciation through anagenesis
Evolution by creeps
Punctuated Equilibrium
Long periods of stasis (no change) → brief periods of rapid morphological change → species by cladogenesis
Evolution by jerks
Diversity
balance between origination and extinction
Turnover
Number of species eliminated and replaced per unit of time
Standing diversity
Number of species present in an area at a given time
Adaptive Radiation
Rapid diversification of a lineage into a range of ecological niche specialists
Origin rate > extinction rate
Ecological Opportunity
Presence of vacant niche space → room for diversification of a lineage into a free niche (role)
Distinct niche specialists favored in different environments → speciation
Absence of predators → ecological opportunity for a lineage
Key Innovation
trait that allows a lineage access to new resources
species take advantage of a resource not previously being used
Evolution novelty
New genetically based trait
Paradox of novelty: changing something that works → worsens life, evolution tinkers and uses old materials in new ways
Novel Gene Generation:
pop. in new env. using accessible enzymes and proteins → slow growth rates since limited resource → fitness improved as pop. grows → pop. large enough → opportunity for modification through selection → fitness improves
Innovation
Act of introducing a novelty
Synonym for evolutionary novelty
Excaptation/Co-option
An existing gene function for one purpose starts preforming a new role
Amplification
Making more of what you have, even if its not the best it possible could be
Paralog
Gene descended from duplication of same genome → have two copies of gene → choose to use one
Divergence
improves fitness through changes in gene function
Amplification and divergence can work together → amplification causes two copies → one diverges while other still keeps original function
Background extinction
normal rate of extinction for taxa or biota
Mass extinctions
statistically significant increase about background extinction rates
What’s the fuel of evolutionary change?
Mutations
Mutations → selection
Recombination only moves things around but doesn’t create new variation
How do mutations affect fitness?
Most are deleterious or neutral, very few are beneficial
What is Fisher’s Geometric Model?
It’s easier to make a well-adapted system worse than better
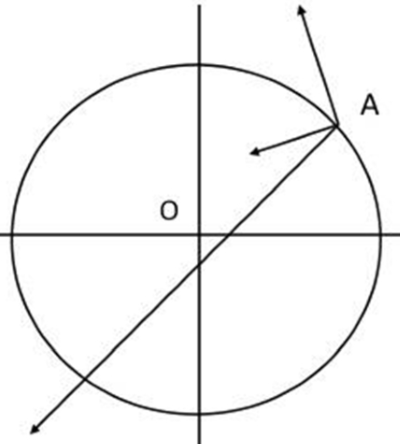
What mutations matter for evolution?
Germ-line mutations (in gametes) are heritable
Somatic mutations (in body cells) are not heritable (ex: cancer)
Why aren’t recombination and independent assortment sources of novelty?
They mix things up and create new genetic combinations but NOT new genetic material or variation
Genotype and Environment Interaction
Phenotype = Genotype + Environment
Genotype
Genetic makeup of an individual
Combo of alleles carried by an individual at a locus of few loci
Phenotype
observable, measurable characteristic
Polyphenic Trait
Single genotype produces multiple phenotypes
Many genes may be involved → express modified by environment
Phenotypic Plasticity
Same gene having multiple phenotypes based on environment
Ploidy
Number of copies of unique chromosomes in a cell
Pseudogenes
Non-functional genes
Types of Mutations
Point, insertion, deletion, gene duplication, inversion, chromosome fusion, genome duplication
Population
Group of interacting and potentially interbreeding individuals of the same species
Genetic locus
Location of specific gene/sequence of DNA on a chromosome
Population Genetics
study of allele distributions and frequencies in space and time
Evolution
Change in allele frequencies form generation to the next
Null Model (in evolution)
Distribution of allele and genotype frequencies if there are no forces acting on it (no selection, drift, mutation, or migration)
Assume null model → no evolution → same frequencies in multiple generations
How do different mutations affect population?
Strongly deleterious: never establish → eliminated very quickly by selection
Beneficial mutations: if not last by chance → sweep very quickly
Mildly deleterious and neutral mutations: can be lost or increase frequency due to chance
Genetic Drift
deviations in allele frequencies caused by chance alone
Bottleneck
Temporary reduction in population size → low genetic diversity/variation (often inbreeding)
Founder Effect
Type of bottleneck
Small # of individuals colonizing a new isolated habitat
Why does natural selection occur?
Replication of hereditary material is imprecise → genetic variation
Some lineages replicate faster
Quantitative Genetics
Evolution of continuous phenotypic traits → polygenic → many genetic loci
More loci → more phenotypes → continuous distributions
Polygenic Trait
trait influenced by many genes at many loci (epistasis) and at one locus (dominance)
Heritability
Proportion of phenotypic variance attributable to genetic differences among individuals → property of population
Ratio of variances
Heritable
Whether something can be inherited or not
Trait can be heritable with no heritability
Stabilizing Selection
Agents of selection oppose each other → average of the two selected for
Ex: detrimental to be too big or too small for separate reasons → average size
Disruptive Selection
Extreme traits favored over average ones
Opposite of stabilizing selection
Leads to speciation
Phylogenetic Species Concept (PSC)
Smallest possible group descending from common ancestor
Recognizable by synapomorphies (unique, derived traits)
Count the nodes
Biological Species Concept (BSC)
Species are groups of potentially interbreeding populations that are reproductively isolated from such other groups
General Lineage Species Concept (GLSC)
Metapopulations that exchange alleles frequently enough to comprise same gene pool (spatially separated populations, any gene flow)
Allopatry
Geographic barrier, spatially separate populations evolve without gene flow between them
Not enough alone to judge if 2 populations are separate species
Reproductive Isolation
Can maintain species barriers even species are together (in sympatry)
RI: Pre-mating
Isolation before mating even occurs
RI: Post-mating/Prezygotic (Copulatory behavioral isolation & Gametic incompatibility)
After mating, before zygote forms
Copulatory behavioral isolation: mating results in damage to female reproductive system
Gametic incompatibility: Sperm fails to fertilize egg
Post-mating/Postzygotic
Hybrids produced often have low fitness, can be sterile
Allopatric Speciation
Geographical isolation → no gene flow
Sympatric Speciation
No geographic isolation → complete gene flow
Ex: plants evolving different flowering times → reproductive isolation
Reinforcement
natural selection favors prezygotic isolation mechanisms → prevent formation of hybrids with reduced fitness
hybrid mating VERY RARE even in sympatry
Ecological Speciation
selection for different ecological traits in different niches → reproductive barriers → pre & post zygotic isolation
Bateson-Dobzhansky-Muler Incompatibility
genetic incompatibility in hybrid offspring caused by epistatic interactions at two or more loci → genes that work fine separately cause problems when together
Allopolyploidy
polyploidy (different number of chromosomes) resulting from interspecific hybridization
extremely rapid selection
AFTER ORIGINAL POLYPLOIDY: entire genome duplication in hybrids of species with different # of chromosomes → viable offspring → often new species
Stable Ecotype Model
Niche specific adaptations define species in microbes