Chapter Sixteen - Mechanisms of Genetic Variation
1/73
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
74 Terms
Mutations
Heritable changes in the DNA sequence that occur in the absence of sexual reproduction. They are characterized by the genotypic change they cause or the change’s phenotypic consequences.
Point Mutations
Mutations that affect only one base pair. These occur through the alteration of single pairs of nucleotides or through the addition or deletion of one nucleotide pair in the coding region of a gene.
Spontaneous Mutations
Mutations that arise occasionally in all cells in the absence of any stimulus.
Induced Mutations
Mutations that are the result of exposure to a mutagen, which can be either a physical or a chemical agent
1) DNA replication errors
2) Collisions between the replisome and RNA polymerase
3) Spontaneously occurring lesions in DNA
4) The actions of mobile genetic elements
What causes spontaneous mutations?
Tautomeric/hydrogen
Replication errors can occur when the nitrogenous base of a nucleotide shifts to its ( ) form rather than its keto form. This causes changes to the ( )-bonding of the bases.
Transition Mutations
Common mutations that result in purine-purine or pyrimidine-pyrimidine substitutions. These are due to changes in the hydrogen-bonding capabilities of nitrogenous bases in nucleotides.
Transversion Mutations
Rare mutations that result in purine-pyrimidine substitutions and vice versa. These cause many steric problems resulting from the pairing of purines to purines and pyrimidines to pyrimidines.
Repeated/AT
Replication errors that result in insertion or deletion of nucleotides typically occur when there is a short stretch of ( ) nucleotides, especially ( ) base pairs.
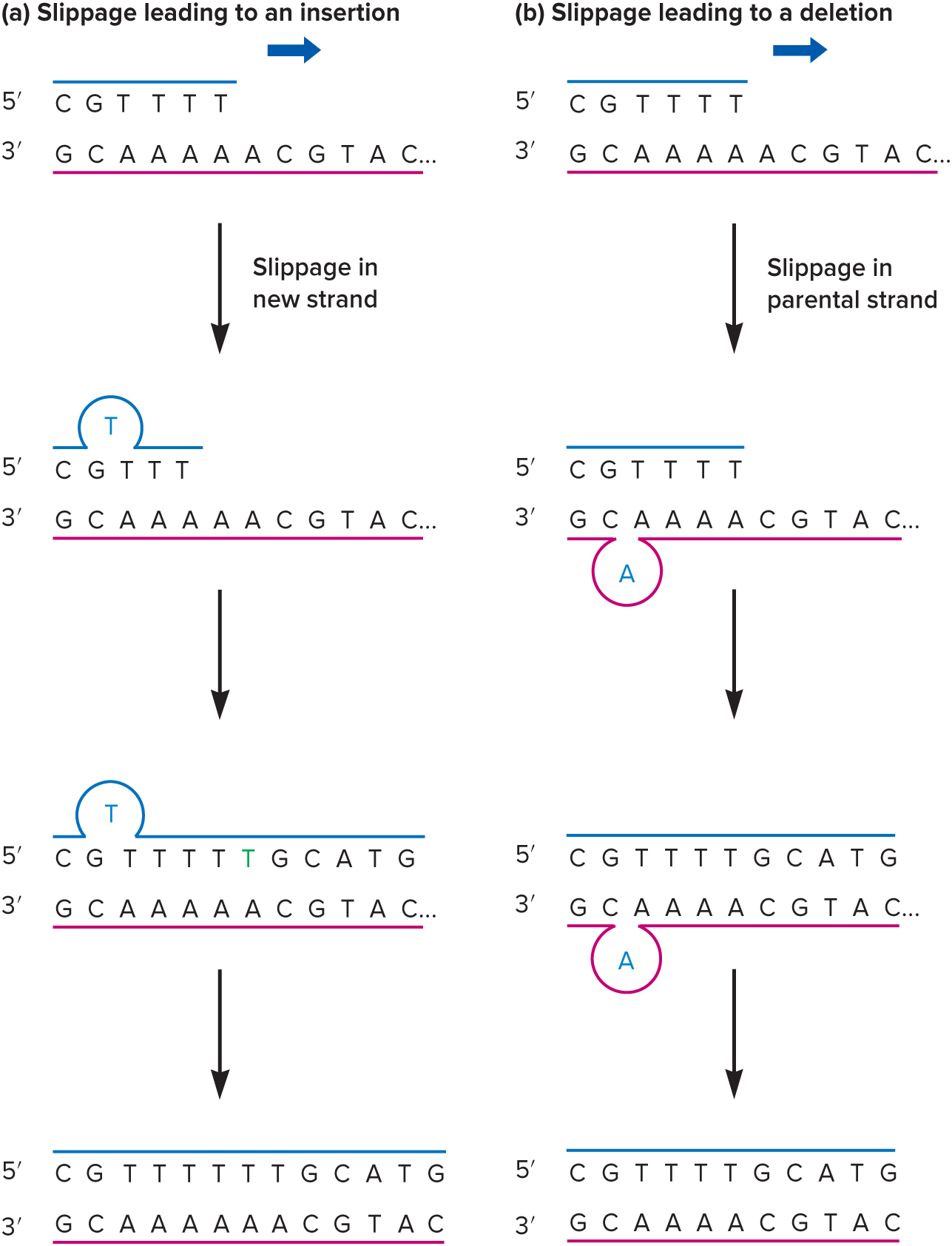
Lesions/apurinic
( ) in DNA can cause spontaneous mutations. For example, purine nucleotides can be depurinated, meaning they lose their base but keep the sugar-phosphate backbone intact. This results in a ( ) site, which can’t base pair and may cause a mutation when the DNA is replicated.
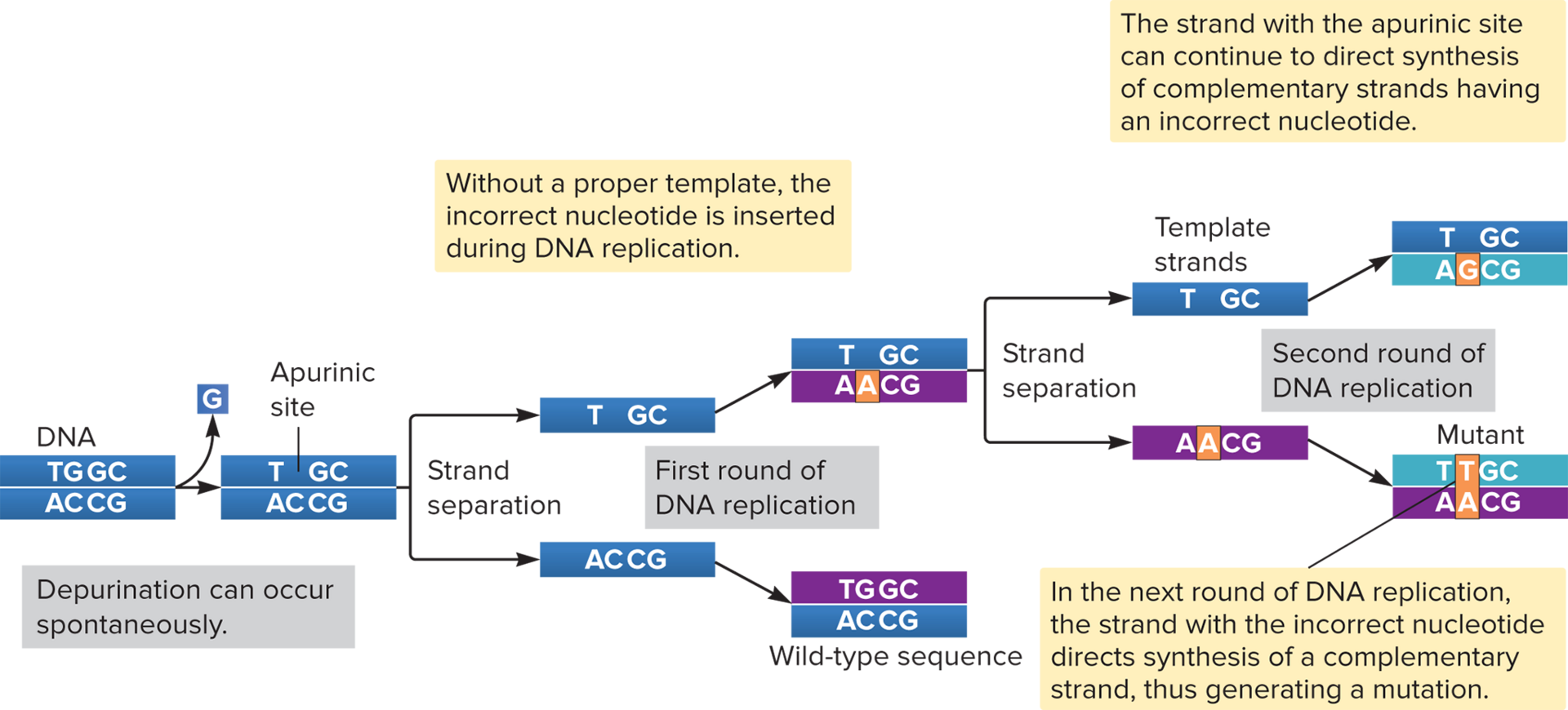
Apyrimidinic Site
This forms when pyrimidine bases are lost from a nucleotide. It can’t base pair and will likely cause a mutation when the DNA is replicated.
1) Base analogues
2) DNA-modifying agents
3) Intercalating agents
What are three common types of chemical mutagens?
Base Analogues
Mutagens that are structurally similar to normal nitrogenous bases and can be incorporated into a growing polynucleotide chain during replication. Once in place, they will exhibit the base-pairing properties different from the bases they replace, causing a stable mutation later on.
Ex) 5-bromouracil undergoes tautomeric shift quite frequently. It replaces thymine and hydrogen bonds like cytosine to guanine rather than adenine.
DNA-Modifying Agents
Mutagens that change a base’s structure and therefore alter its base-pairing specificity. They often react preferably with specific bases and cause a particular type of DNA damage.
Intercalating Agents
Mutagens that distort DNA to induce single nucleotide pair insertions and deletions. Their planar structure allows them to insert themselves between the stacked bases of the helix, distorting it and rendering the DNA inactive.
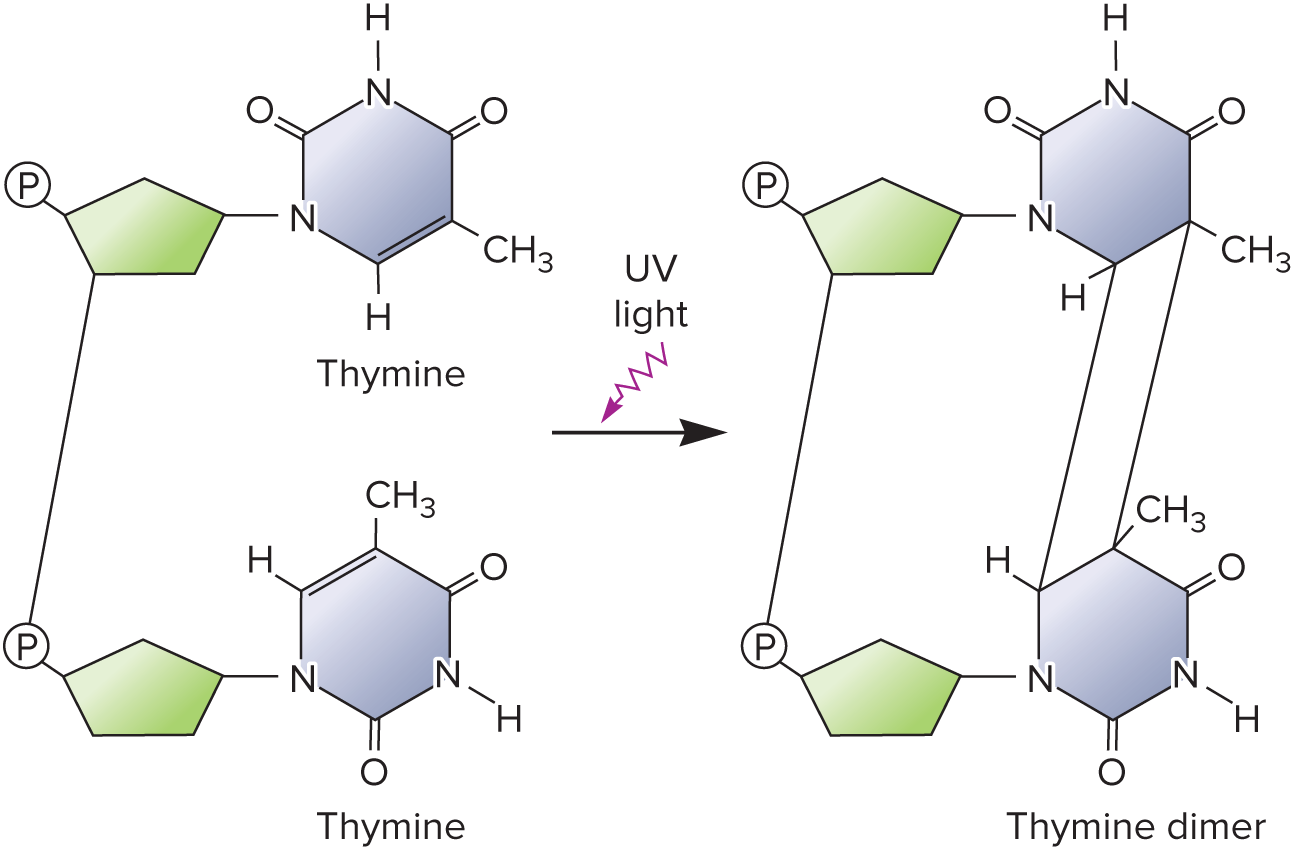
Thymine dimers
Ultraviolet radiation will generate ( ) between two thymine bases on the same strand, irreversibly impairing the DNA strand and making it unable to replicate.
Wild-Type
Used to describe the more prevalent form of a gene and its associate phenotype.
Forward Mutation
A mutation that transforms a wild type gene to its mutant form.
Silent Mutations
Mutations that change the nucleotide sequence of a codon but do not change the amino acid encoded by that codon. This is possible because the genetic code exhibits degeneracy and each codon codes for more than one amino acid.
Missense Mutations
Mutations that involve a single base substitution that changes a codon for one amino acid into a codon for a different amino acid. The effects of these mutations depends on the type and location of the amino acid substitution.
Neutral Mutations
These occur when missense mutation has little or no effect on the phenotype due to the type or location of the amino acid substitution.
Ex) The replacement of one polar amino acid for another polar amino acid at the protein surface as a result of missense mutation will have little or no effect on the protein.
Nonsense Mutations
Mutations that convert a sense codon, which is one that codes for an amino acid, to a nonsense codon, or stop codon, and causes early termination of translation and results in a shortened polypeptide.
These mutations cause complete loss of function when they occur closer to the beginning or middle of the gene.
Frameshift Mutations
Mutations that arise from the insertion or deletion of base pairs within the coding region of the gene. Since the code consists of a precise sequence of codons, the addition or deletion of one or two base pairs causes the reading frame to be shifted for all subsequent codons.
Reversion Mutation
A second mutation that occurs at the same site as the original mutation and restores the wild-type phenotype by either restoring the original sequence or by creating a different codon for the same amino acid.
Suppressor Mutation
A second mutation that occurs at a different site than the original mutation and restores the wild-type phenotype. It is classified as either intragenic or extragenic depending on whether it occurred in the same gene or a different gene, respectively.
Conditional Mutations
A type of lethal mutation in which the mutation expresses a different phenotype only under certain environmental conditions.
Ex) A mutation in a bacterium might not be expressed when the bacterium is grown in low temperatures but is when grown in high temperatures.
Auxotrophs
Mutated organisms with a conditional phenotype, meaning they are unable to grow on a medium lacking a specific molecule required for their survival.
Prototroph
The wild-type organism from which the mutant auxotroph arose. They are able to grow on minimal media containing only salts and a carbon source.
Low
tRNA- and rRNA-encoding genes exhibit a ( ) frequency of mutations compared to protein-encoding genes because of their critical role in RNA structure and protein synthesis.
Physiological Suppression
A type of extragenic suppression that occurs when a defect in one pathway is circumvented by a second mutation that induces expression of another pathway to the same product or by a mutation that permits more efficient uptake of a compound affected by the first mutation.
Selective pressure
( ) determines if a mutation will persist in a population to become an alternate form of the gene, termed an allele. ( ) simply refers to the idea that organisms in a population will each have a unique genotype that produces a unique phenotype favored under different environmental condition because they lead to increased survival.
Recombination
The process in which one or more nucleic acid molecules are rearranged or combined to produce a new genotype. Organisms produced following this process are referred to as recombinants.
Vertical Gene Transfer
The transfer of genes from sexually-reproducing parents to progeny. This process produces genetic recombination in two ways: crossing-over between sister chromatids during meiosis and the fusion of gametes to produce a viable zygote.
Horizontal (Lateral) Gene Transfer
A process of genetic recombination in bacteria and Archaea that transfers genes from one independent, mature organism to another mature organism, creating a stable recombinant having characteristics of both donor and recipient.
It commonly occurs between microbes sharing a habitat and helps bacteria and archaean survive environmental stresses like UV damage and antibiotics.
1) Transformation
2) Conjugation
3) Transduction
What are three mechanisms of horizontal gene transfer?
1) If the donor DNA has a sequence homologous to the recipient DNA, it may integrate and recombine with the host’s DNA
2) If the donor DNA can replicate apart from the recipient (i.e. it is a plasmid), it may exist and replicate separately from the recipient DNA when the recipient reproduces
3) The donor DNA remains in the cytoplasm and doesn’t replicate
4) The donor DNA is degraded or restricted by the recipient
What can happen to donor DNA once it enters a recipient during horizontal gene transfer?
Homologous Recombination
The most common mechanism for crossing-over in all organisms. It occurs when long regions of the same or similar nucleotide sequence in two different DNA molecules break and reunite in a crossing-over pattern.
Ex) Similar sequences on a chromosome and a plasmid will undergo crossing-over in this way.
Site-Specific Recombination
A major type of recombination that differs from homologous recombination in that it does not require similar sequences, it occurs at specific target sites in the DNA molecules, and it is catalyzed be specific enzymes called recombinases. It is commonly used by plasmids and viral genomes to integrate into host chromosomes.
Transposable Elements
Also called mobile genetic elements, these are genetic elements that move within and between genomes.
Transposition
The movement of genetic elements between genomes or within genomes.
Transposase
The recombinases used by a specific mobile genetic element may be called an integrate, resolvase, or ( ).
Insertion Sequences (IS Elements)
Short sequences of DNA that typically contain the gene for transposase and are bounded by inverted repeats of identical or similar sequences of nucleotides in reverse orientation. Transposase will recognize the ends of this structure and catalyze transposition.
Transposons
These consist of a central region containing genes unrelated to transposition flanked by IS Elements that are identical or very similar in sequence. The flanking IS elements encode the transposase that enables this structure to move and contain strong promoters that activate nearby genes.
Simple Transposition
Also called cut-and-paste transposition, this process of transposition involves transposase catalyzing excision of the MGE followed by cleavage of a new target site and ligation of the element into this site. The transposase also cleaves the target site, leaving 5 to 9 base pair overhangs that can later hold direct repeats and inverted repeats.
Replicative Transposition
In this type of transposition, the MGE remains at the original site and a copy is inserted at the target DNA site.
Conjugation
The transfer of DNA by direct cell-to-cell contact.
Plasmids
Small, double-stranded DNA molecules that can exist independently of host chromosomes. They have their own replication origins, replicate autonomously, and are stably inherited.
F Factor
A conjugative plasmid roughly 100,000 base pairs in length that bears the genes responsible for cell attachment and plasmid transfer between E.coli cells. Because it contains several IS elements that mediate plasmid recombination with the host cell chromosome, it is said to be an episome that can exist outside the chromosome or be integrated into it.
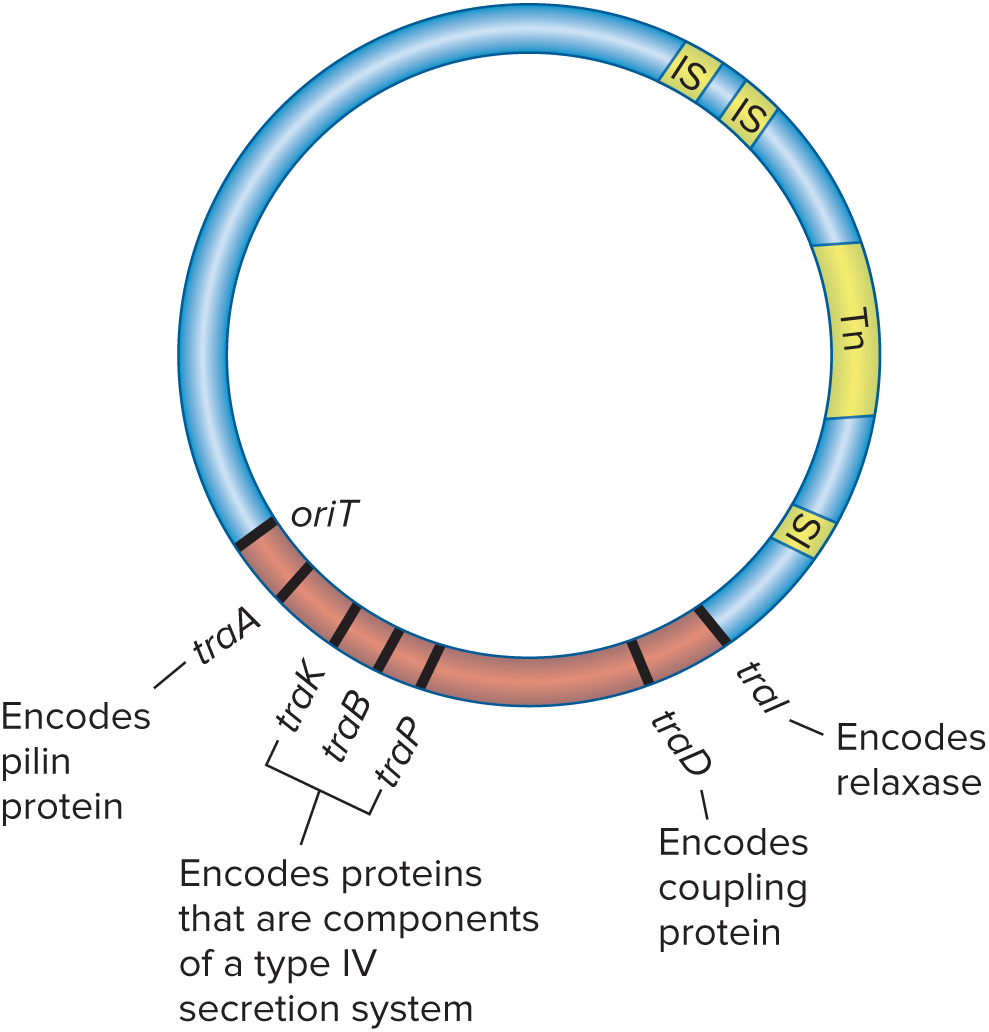
tra operon
Much of the information needed for plasmid transfer in the F factor is located in the ( ), which contains genes that direct the formation of the sex pilus and others that aid in DNA transfer.
F+ Cell
The donor cell that contains the F plasmid.
F- Cell
The recipient cell that does not contain the F plasmid but instead received DNA from the F plasmid of a donor cell.
Lederberg & Tatum
These two took two auxotrophic strains, incubated them in a mixture for several hours, and then plated the mixture on minimal medium. They used double and triple auxotrophs to ensure reversion or suppressor mutations were rare. They found that recombinant prototrophic colonies appeared and concluded that the two auxotrophic colonies associate and underwent recombination.
Davis
Found that when two auxotrophic strains were separated from each other by a gass filter, which allowed the passage of media but not cells, the strains did not undergo gene transfer. He concluded that direct contact was needed for the cells to undergo recombination, which he deemed conjugation.
Hayes
Rarely/Unidirectional/F+
Found that in F+ x F- mating, the progeny were ( ) changed with regard to auxotrophy, the process was ( ), and that F- strains often became ( )>
Sex Pilus
A cell appendage used to establish contact between the F+ and F- cells.
1) The sex pilus, once produced through expression of the F+ plasmid, established contact between the F+ and the F- cell
2) The pilus retracts, pulling the F- cell towards the F+ cell
3) A type IV secretion system termed T4SS is produced by the F+ cell to join the two cells with proteins from the sex pilus
4) Replication enzymes cleave the F+ plasmid and begin its replication via the rolling-circle mechanism
5) The TraI protein guides the displaced strand from step four through the T4SS to the recipient cell
6) The strand entering through the T4SS is used as a template strand and replicated, converting the F- species into F+
What are the steps of F+ x F- conjugation?
Rolling-Circle Replication
A process that replicates DNA during conjugation. It starts with relaxosome, an enzyme that cuts the circular DNA. As a result, the 3’ end is freed and extended with replication enzymes while the 5’ end is displaced to form an ever-lengthening tail.
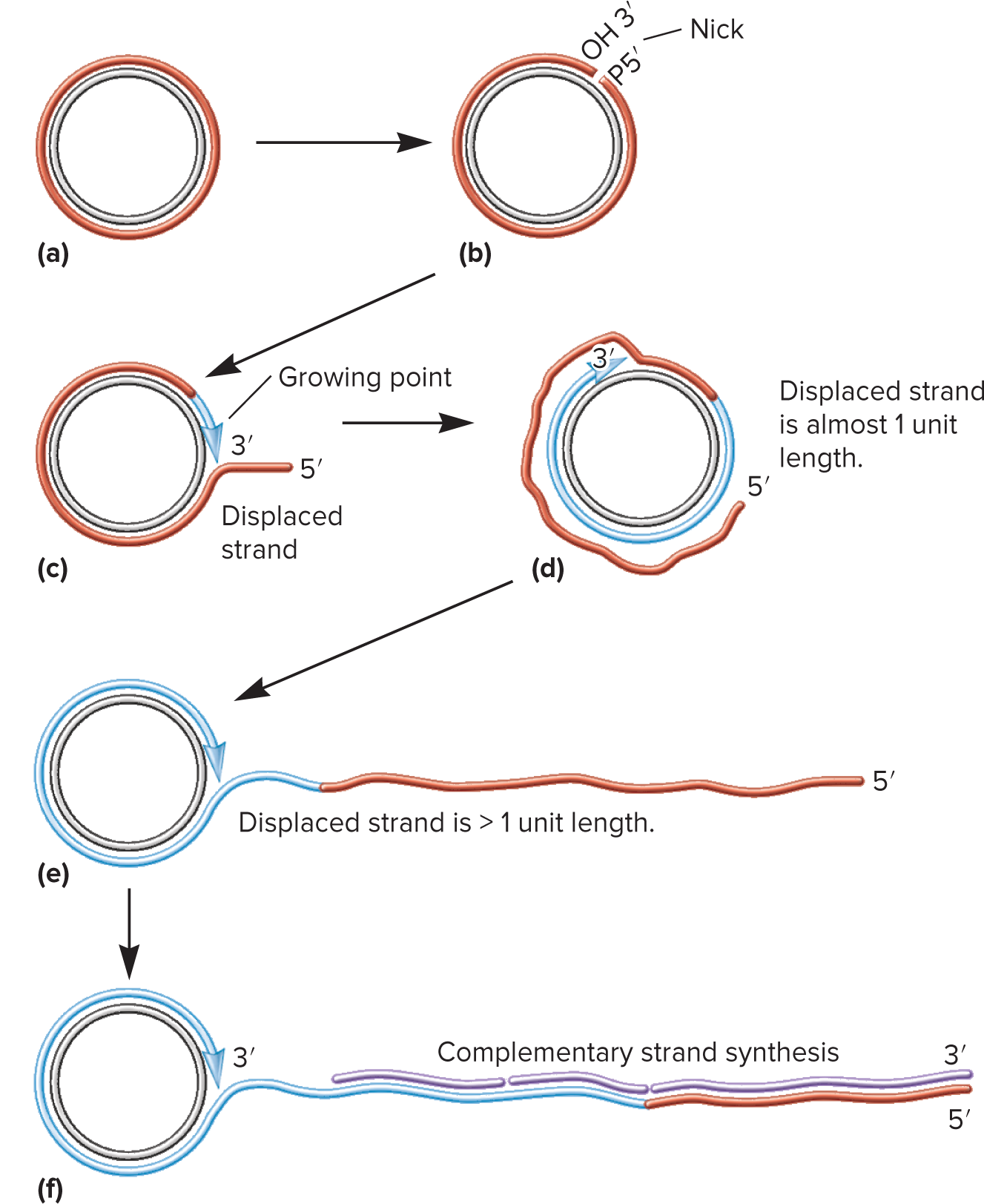
Relaxosome
A complex of proteins encoded by the F factor that nicks the conjugate plasmid at the origin of transfer (oriT) site. A major component of this complex is a protein called TraI, which remains attached to the displaced 5’ end and guides it through the T4SS.
Hfr Conjugation
A type of conjugation that transfers chromosomal genes with great efficiency but does not change the recipients into F+ cells. This type of conjugation occurs when F plasmids are rarely integrated or recombined into the main chromosome. It is called this because of the high frequency of recombinants produced.
F-
Hfr conjugation begins just like F conjugation, but rather than just transferring the F plasmid to the recipient, it also directs the transfer of the host chromosome. If given the full 100 minutes to do so, the whole host chromosome is funneled through the recipient and replicated. However, this rarely occurs, and only part of the F factor is transferred. Because of this, the recipient remains ( ).
F’ Plasmid
An F plasmid that tried to excise itself from the bacterial chromosome but ended up excising a part of the chromosome and taking it with it. Thus, this species is an F plasmid containing a bit of the host chromosome and is genotypically distinct and larger than the original F factor.
F’ Conjugation
In this type of conjugation, the F’ plasmid is fed through and replicated in the recipient cell, converting the recipient into an F’ cell. In this process, only the genes on the F’ plasmid are transferred, just as with F conjugation. The genes transferred don’t need to be incorporated into the recipient chromosome to be expressed.
Transformation
The uptake of circular or linear DNA from the environment outside the cell and maintenance of the DNA in the recipient cell in a heritable form. It occurs naturally when bacteria lyse and release their DNA into the surrounding environment.
Competent Cell
A cell that is able to take up DNA and be transformed - the DNA is bound to the cell and imported if the fragment comes in contact with this.
Similar/type II/pili
Gram-positive and Gram-negative bacteria use ( ) proteins as the main components of their DNA uptake machinery. These proteins are related to those used in ( ) secretion systems and type IV ( ).
ComEA/ComEC
During transformation in S.pneumonie, the DNA uptake pilus retracts upon contacting DNA and brings it to the plasma membrane surface. A membrane-bound protein ( ) directs the DNA to the membrane channel ( ) and an endonuclease (N). The one strand passes through the channel while the other is degraded.
Transduction
A mode of horizontal gene transfer mediated by viruses like bacteriophages. It involves the transfer of bacterial or archaeal genes by virus particles.
Virulent Bacteriophages
Bacteriophages that multiply in their host immediately after entry. After the progeny phage particles reach a certain number, they cause the host to lyse, so they are released to infect new host cells.
Lytic Cycle
The process by which progeny phage particles, shortly after their synthesis, lyse their host cell and are released into the environment to infect other cells.
Temperate Bacteriophages
Bacteriophages that establish a relationship with their host called lysogeny, in which they insert their genomes into the bacterial chromosome.
Prophage
The inserted viral genome of a lysogenic relationship.
Generalized Transduction
A type of transduction that occurs during the lytic cycle of virulent phages and sometimes during the lytic cycle of temperate phages. It occurs when the virus begins to take control of its host and degrade the host genome. Occasionally, a fragment of the host genome that is about the same size as the phage genome can be mistakenly packaged into a virion during virion assembly. Once released, this phage containing the host genome may encounter a susceptible host and eject bacterial DNA into that new cell.
Since it lacks viral genes, it can’t initiate a lytic cycle in its new host.
Transducing Particle
A phage that mistakenly carries DNA from its host’s genome and transfers it to another bacterial cell.
Specialized Transduction
A type of transduction in which only specific portions of the bacterial genome are carried by transducing particles and transferred to another bacterial cell. It occurs because of errors during the lysogenic life cycle of temperate phages; if a prophage is excised improperly from the host chromosomes when it comes time to leave, it may carry a specific portion of the host’s genome away along with the virulent DNA.