week 4 pt 2 - unsupervised learning
1/19
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
20 Terms
supervised learning
labelled observations - pairs of data points, each pair consists of a feature vector (x) + its corresponding output label (y) which are related according to an unknown function f(x) = y
during training- use these pairs to learn the relationship between x+y i.e. find a function (or model) h(x) that best fits the observations
goal = ensure that the learned model h(x) accurately predicts the output label of a previously unseen, test feature input (generalisation)
labels = ‘teachers’ during training + ‘validator’ of results during testing
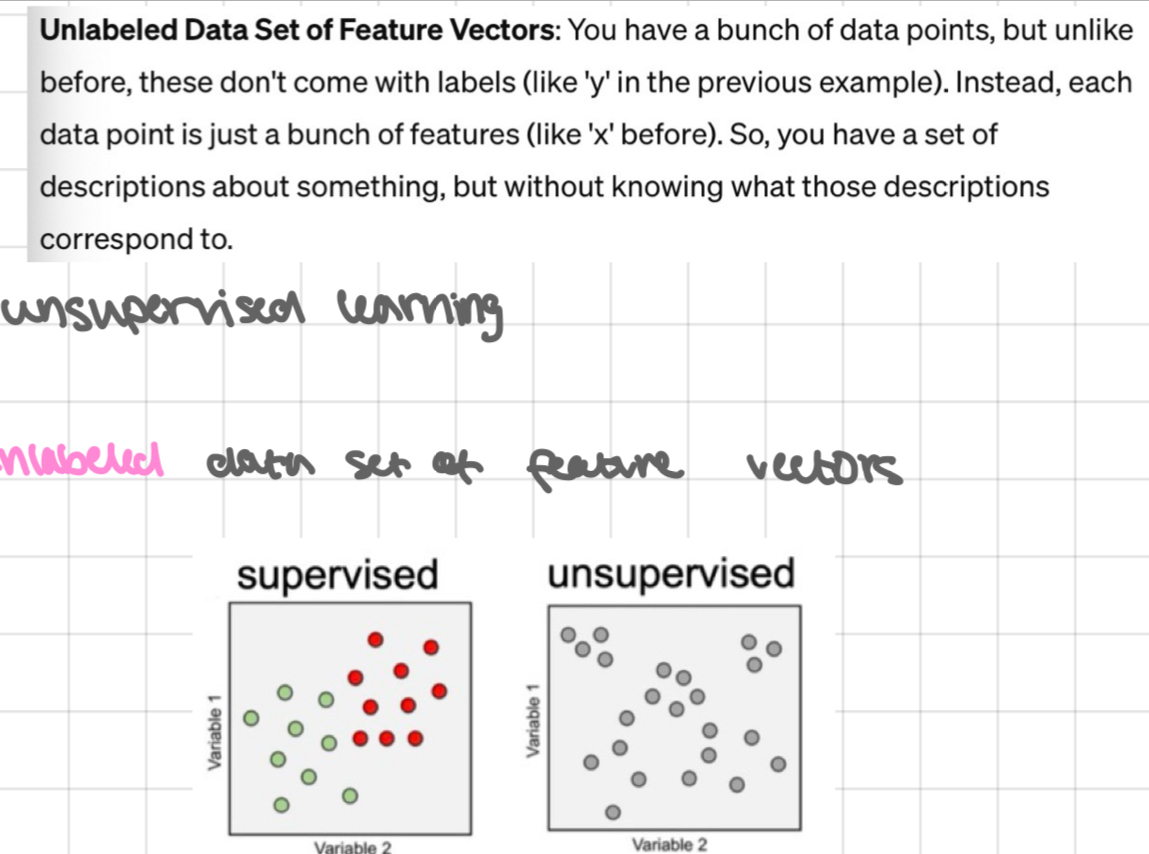
unsupervised learning
unlabelled data set of feature vectors
what can we deduce?
even without labels, can still learn some interesting stuff from this data:
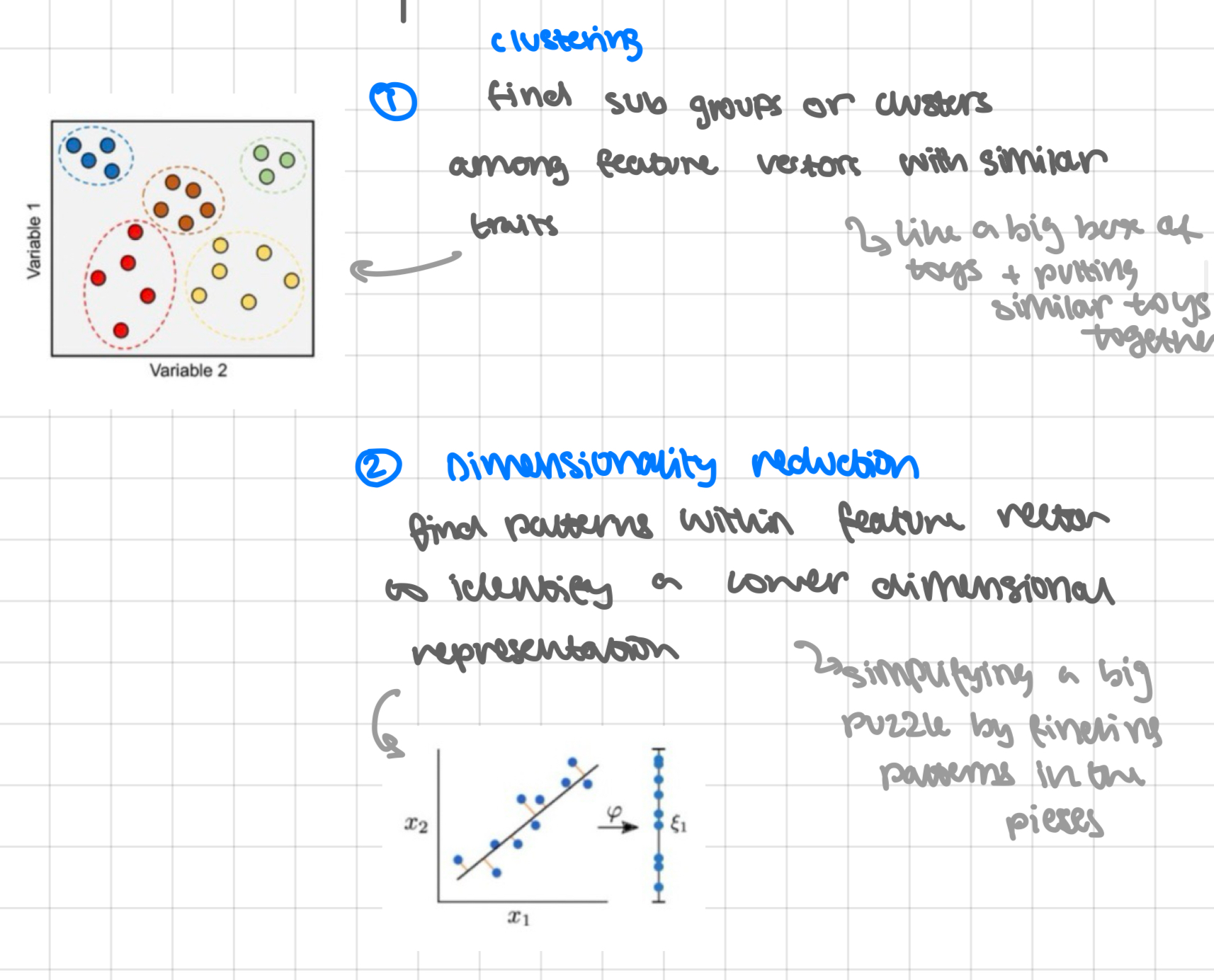
clustering find sub groups or clusters among feature vectors with similar traits
like a big box of toys + putting similar toys together
dimensionality reduction find patterns within feature vector to identify a lower dimensional representation
simplifying a big puzzle by finding patters on the pieces
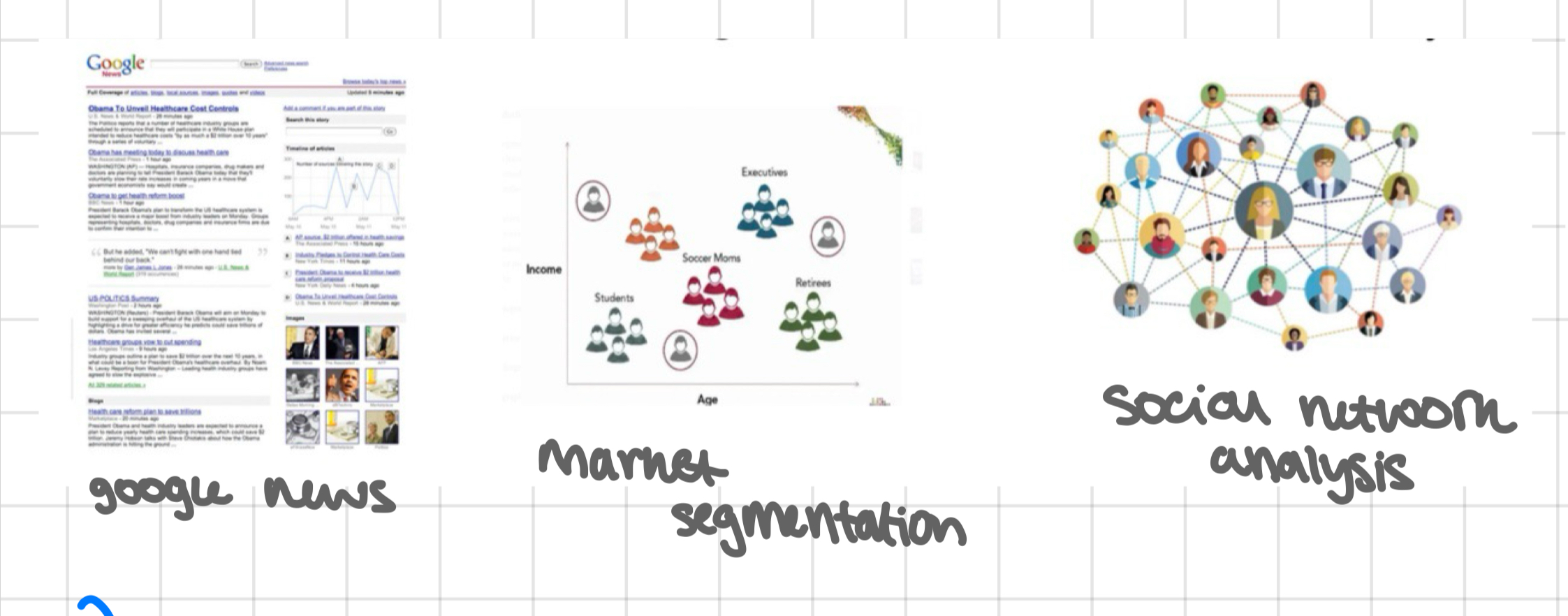
CLUSTERING - real world applications
google news
market segmentation
social network analysis
clusters = potential ‘classes’; clustering algorithms automatically find ‘classes’
DIMENSIONALITY REDUCTION - application
image compression
techniques:
principal component analysis - finds main themes in data
non negative matrix factorisation - breaks down complex mixtures
linear discriminant analysis - helps you desperate diff groups based on their features

why unsupervised learning?
labelled data = £££ + difficult to collect - unlabelled = cheap + abundant
compressed representation - saves on storage + computation
reduce noise, irrelevant attributes in high dimensional data
pre processing step for supervised learning
often used more in exploratory data analysis
challenges in unsupervised learning
no simple goal as in supervised learning
validation of results is subjective
fundamentals of clustering
goal: find natural groupings among observations/objects/feature vectors
segment observations into clusters/groups such that:
objects within a cluster have high similarity (high intra-cluster similarity)
objects across a cluster have low similarity (low intra-cluster similarity)
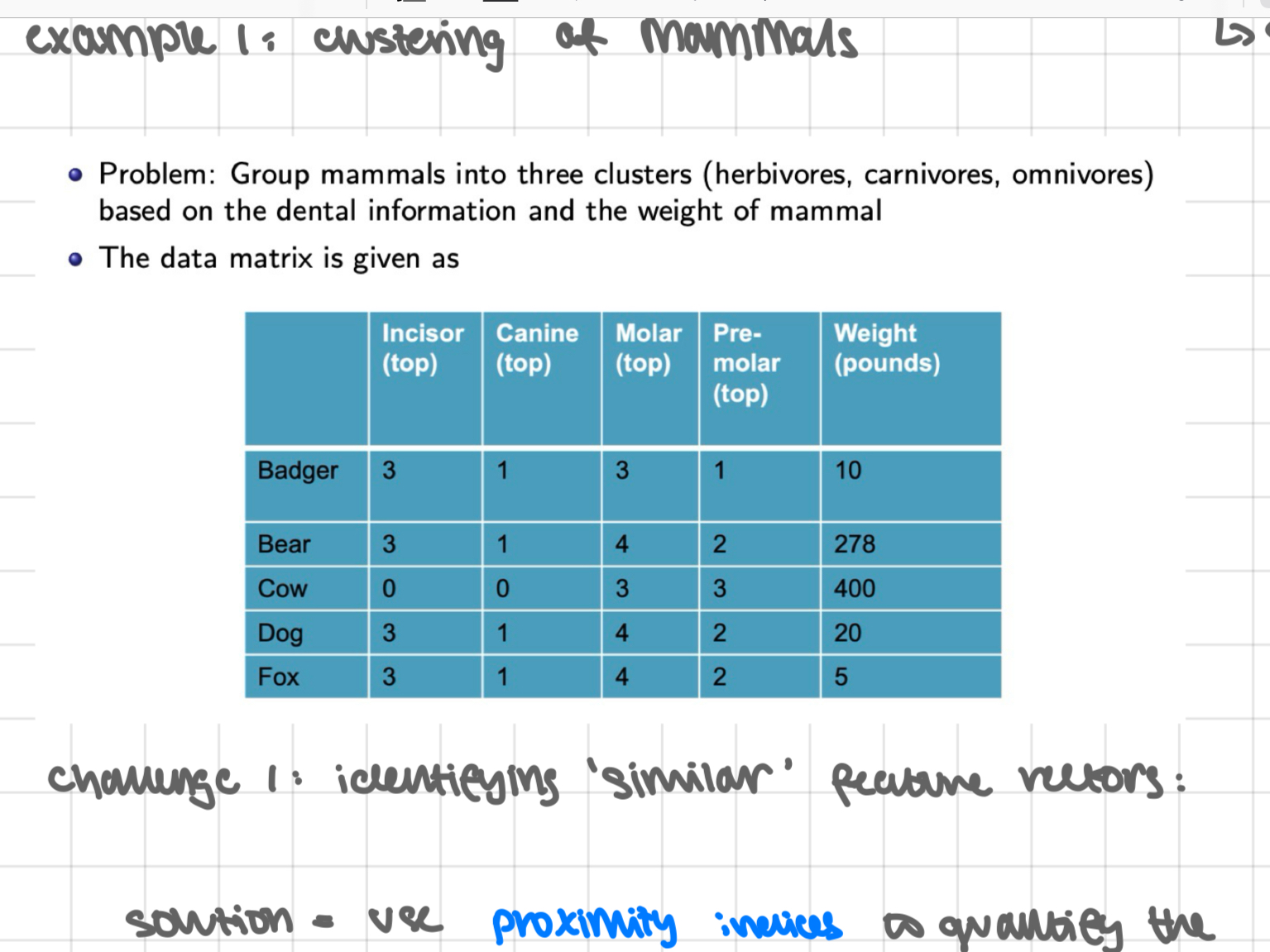
example 1: clustering of mammals
challenge 1: identifying ‘similar’ feature vectors:
solution = use proximity indices to quantify the strength of a relationship between any 2 feature vectors
depends on data type:
continuous valued feature attributes
e.g. x = (0.1, 11.65, 15, 1.4)
nominal feature attributes
e.g. x = (‘large’, ‘medium’, ‘small’)
must be mapped to discrete values consistently e.g. ‘large’ → 3, ‘medium’ → 2, ‘small’ → 1
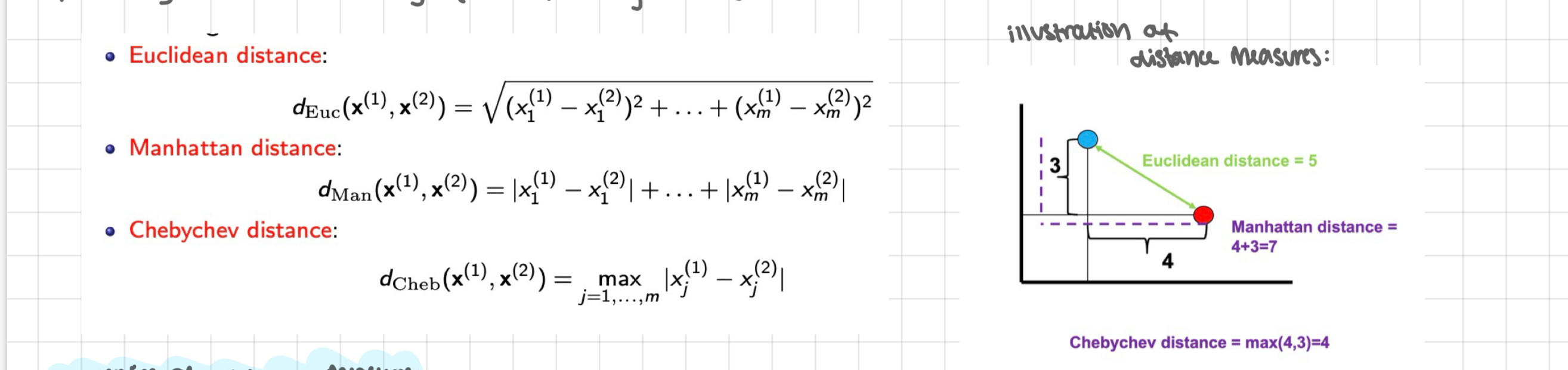
continued
continuous valued features: for 2 continuous valued features x(1) = (x(1), …, x(1)m) and x(2) = (x(2), …, x(2)m), the proximity index can be any of the following distances
euclidean distance
Manhattan distance
chebychev distance
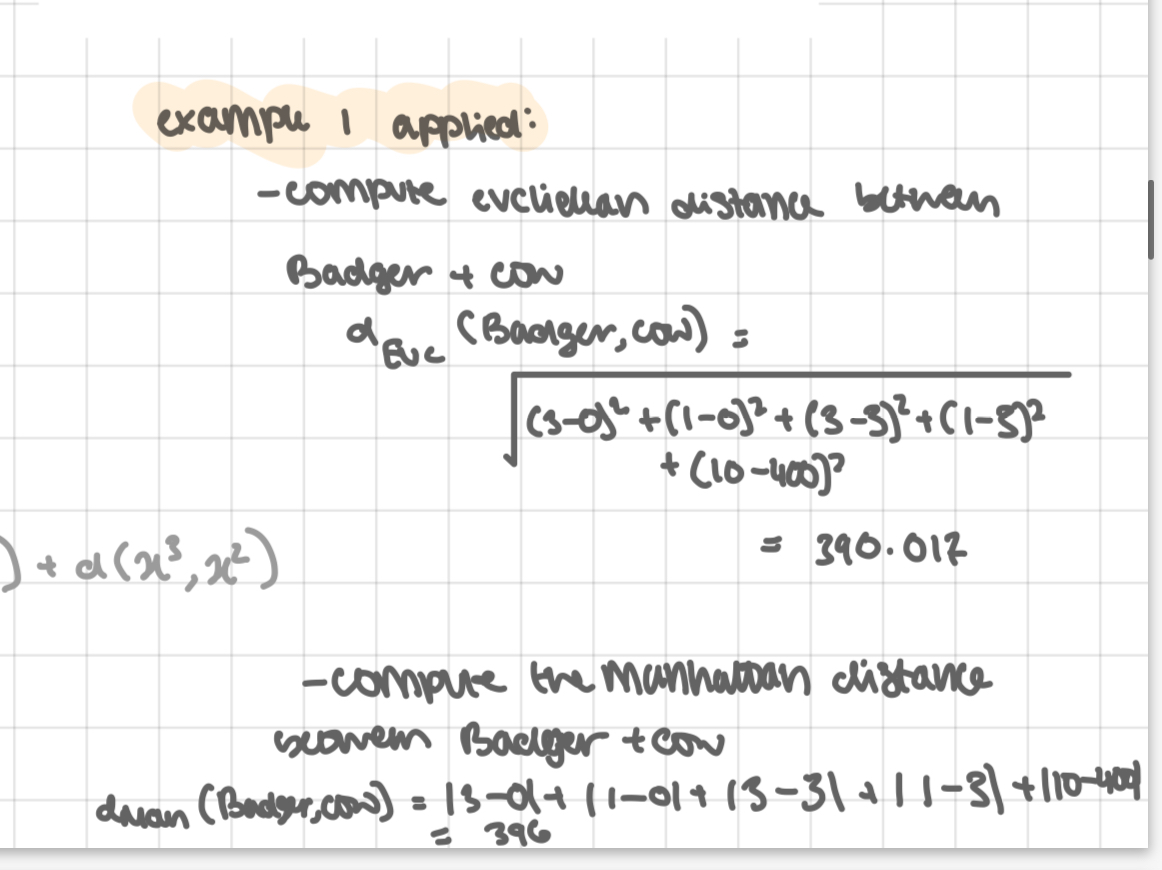
example 1 applied: see pic
continued pt 2
properties of distance measures:
distance between 2 points is always non negative
distance between a point to itself is 0
distance measure is symmetric
distance measure satisfies a triangle inequality
main takeaways:
- diff choice of distance functions yields diff measures of similarity
- distance measures implicitly assign more weighting to features with large ranges than those with small ranges
- rule of thumb: when x prior domain knowledge → clustering should follow the principle of equal weighting to each attribute
this necessitates feature scaling of vectors

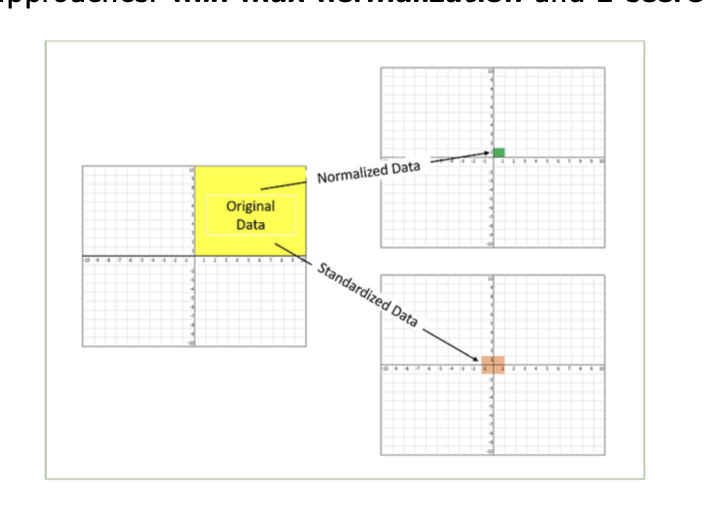
challenge 2: feature scaling
when feature attributes have large differences in the range of values, scaling of features = essential - so that each attribute contributes equally to the distance feature
2 well studied approaches:
min-max normalisation
z score standardisation
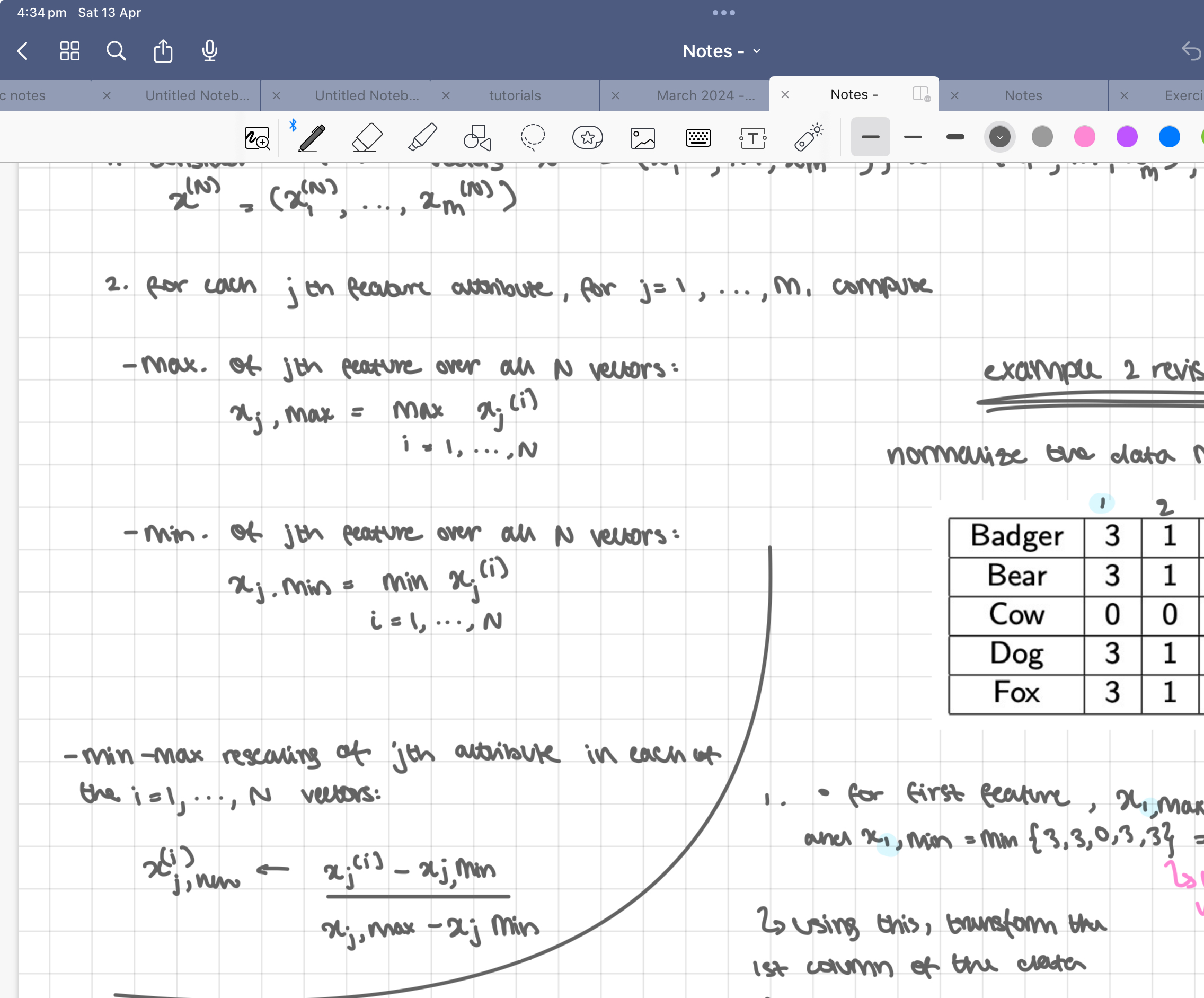
minimum maximum normalisation
all feature attributes are rescaled to lie in the [0, 1] range
steps:
consider N feature vectors x(1) = (x(1), …, x(1)m), x(2) = (x(2), …, x(2)m), x(N) = (x(N), …, x(N)m)
for each jth feature attribute, for j = 1, … m, compute
max. of jth feature over all N vectors:
xj,max = max xj(i) i = 1,…,N
min. of jth feature over all N vectors:
xj,min = min xj(i) i = 1,…,N
min-max rescaling of jth attribute in each of the i = 1, …, N vectors:
x(i)j, new = (xj(i) - xj(i)min)/ (xj(i)max - xj(i)min)

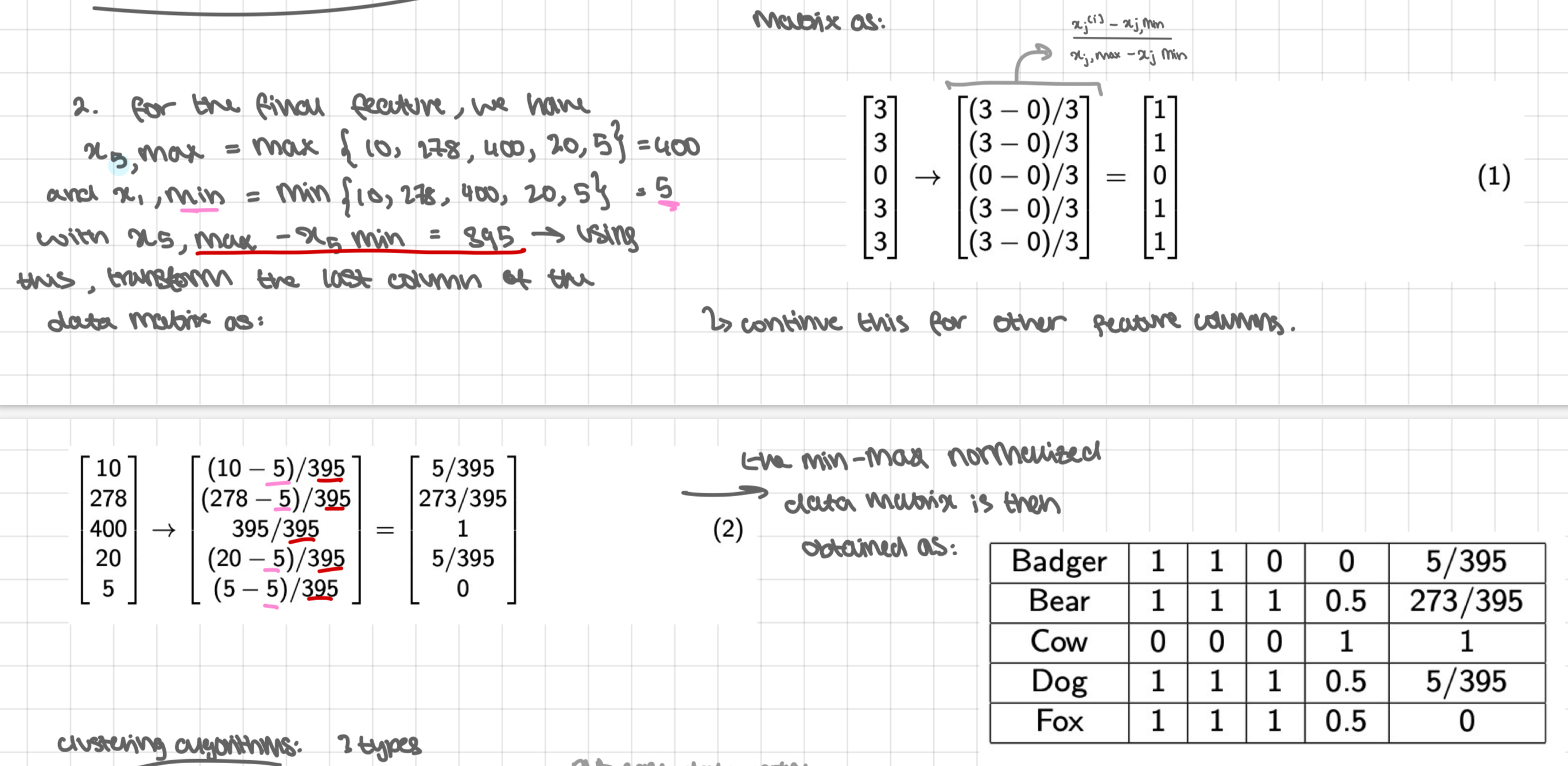
example 2 revisited: normalisation
CLUSTERING ALGORITHMS: 3 types
→ PARTITIONAL CLUSTERING ALGORITHMS
generate a single partition of the data to recover natural clusters
input: feature vectors
examples: k-means, k-medoids
put each data point into 1 group or another based on certain characteristics they share
→ HIERACHICAL CLUSTERING ALGORITHMS
generates a sequence of nested partitions
input: distance matrix, which summarises the distances between diff feature vectors
example: agglomerative clustering, divisive clustering
instead of just 1 partition, they generate a sequence of nested partitions = organise data points into clusters within clusters
→ MODEL-BASED CLUSTERING ALGORITHMS
assumes that the data is generated independently + identically distributed from a mixture of distributions, each of which determines a diff cluster
example: Expectation-Maximisation (EM)
partitional clustering algorithms: the basics
goal: assign N observations into k clusters, where k<N that ensure high intra cluster similarity + low cluster similarity
high = clusters are v close
low = clusters are v. far
can be formulated as a combinational optimisation problem* = requires defining a measure of intra-cluster similarity
where we search for the best arrangement of clusters among all possible arrangements
Notation:
C denote a clustering structure with K clusters
c ∈ C denote a component cluster in the clustering structure C
e ∈ c denote an example in cluster c

measuring intra-cluster similarity
like a ruler to see how close or similar things are within a cluster
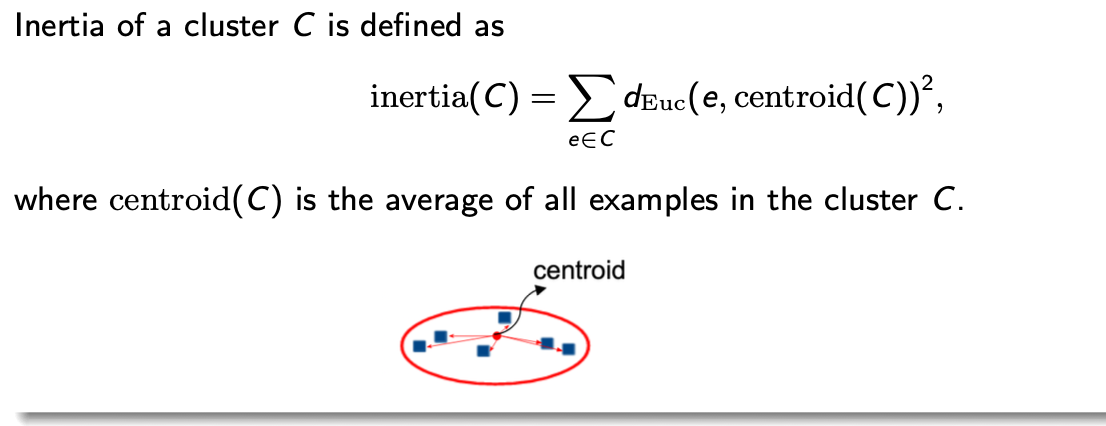
inertia of a cluster c is defined as
inertia(c) = Σe ∈ c deuc (e, centroid(c))²
where centroid (c) is the average of all examples in the cluster c
inertia determines how compact the cluster is. Smaller the inertia, more similar the examples are within the cluster
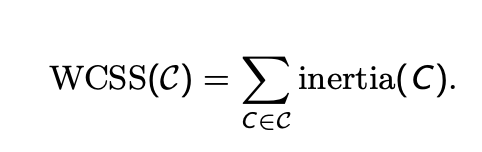
within cluster sum of squares (wcss)
of a clustering structure c consisting of k clusters is defined as
wcss(c) = Σe ∈ c inertia(c)
the sum of the inertia of all clusters in the clustering structure
this quantifies the inertial of all clusters in the clustering structure
used to evaluate the compactness of clusters in clustering algorithms - measures total inertia/spread of all clusters in the clustering solution
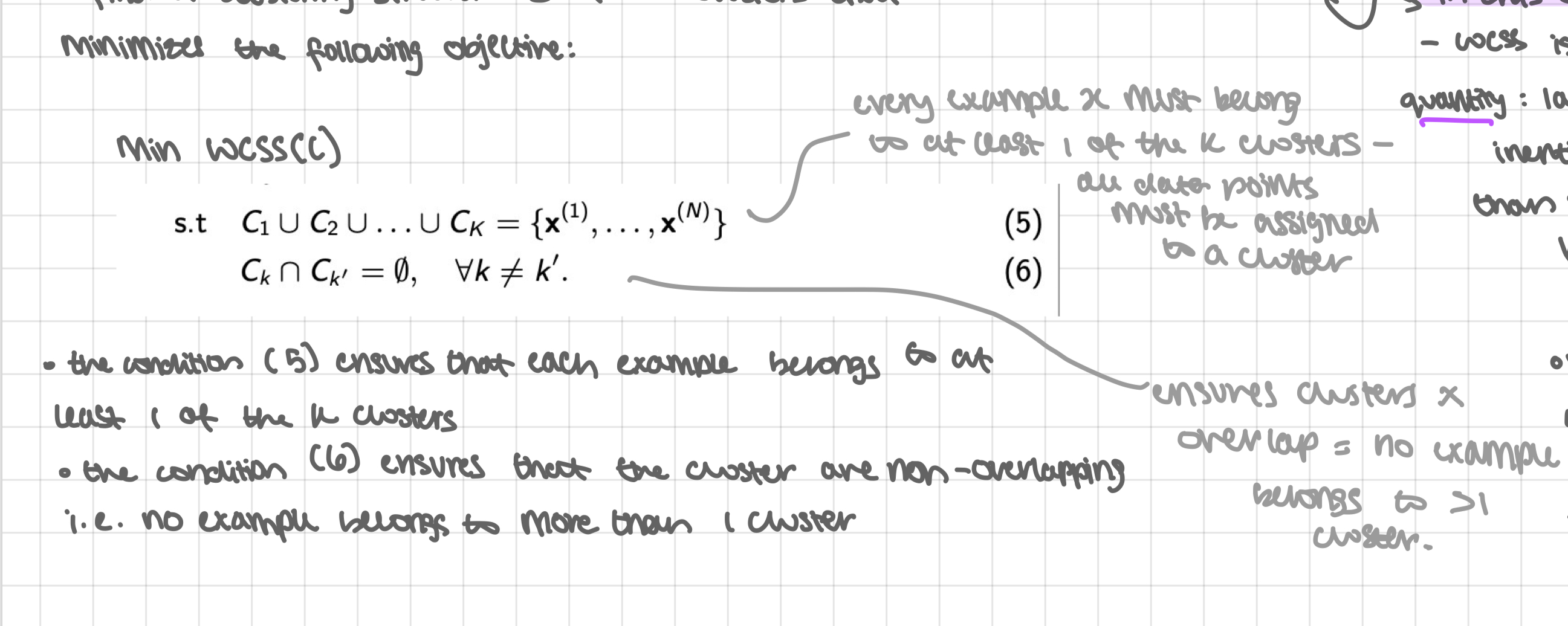
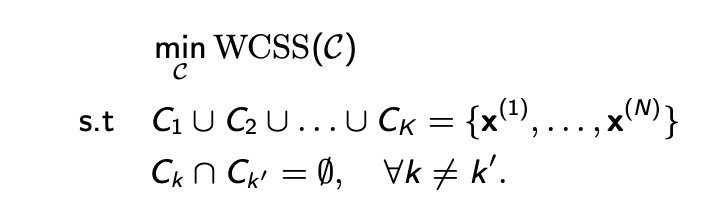
optimisation problem
find a clustering structure c of K clusters that minimises the following objective: (see pic)
the condition (5) ensures that each example belongs to at least 1 of the k clusters
the condition (6) ensures that the clusters are non-overlapping i.e. no example belongs to more than 1 cluster
in this optimisation problem:
wcss is an unnormalised quantity: larger clusters with high inertia are penalised more than smaller clusters with high inertia
minimising wcss of a clustering structure equivalently maximises the intra-cluster similarity
solving the optimisation problem
- finding exact solution of above problem = prohibitively hard
there are almost kN ways to partition N observations into k clusters
infeasible when N is large
hence → look for suboptimal approximate solutions via iterative algorithms
- includes algorithms like k-means, k-medoids
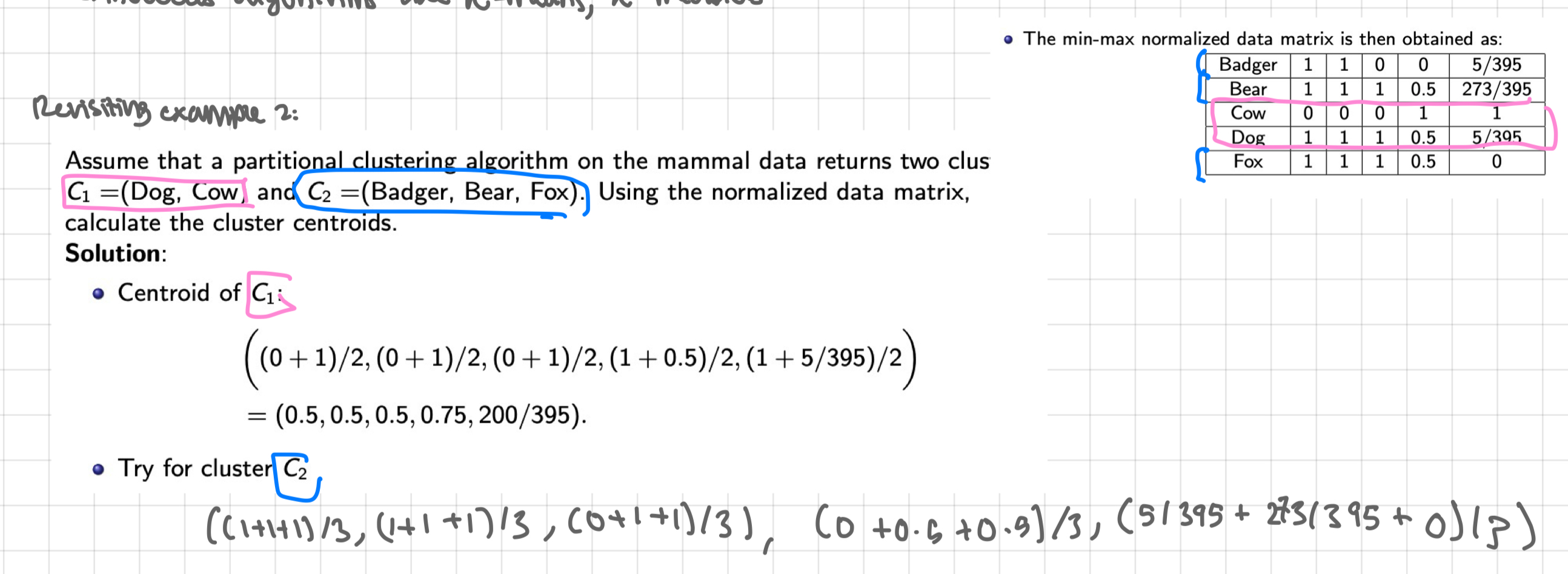
revisiting example 2
assume that a partition clustering algorithm on the mammal data returns two clusters C1= (Dog, Cow) and C2= (Badger, Bear, Fox). Using the normalised data matrix, calculate the cluster centroids