genetics topic 13
1/11
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
12 Terms
DNA microarray purpose
determine the genotype at an SNP
Genomic resequencing purpose
to identify SNPs and CNVs
RNA microarray purpose
To measure level of gene expression
RNA sequencing purpose
to measure amount of transcription of genes expressed
Does DNA microarray look at:
level of gene expression
alternate spicing
CNVs
SNPs
SNPS
Does RNA microarray look at:
level of gene expression
alternate spicing
CNVs
SNPs
level of gene expression
Does genomic resequencing look at:
level of gene expression
alternate spicing
CNVs
SNPs
CNVs and SNPs
Does RNA sequencing look at:
level of gene expression
alternate spicing
CNVs
SNPs
level of gene expression, alternative splicing, and SNPs
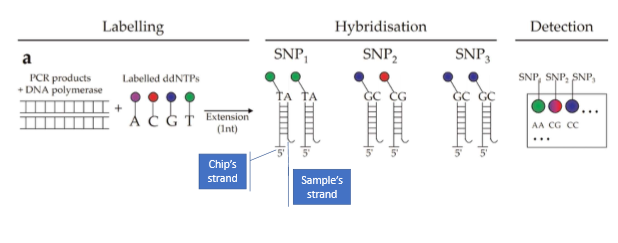
How does DNA microarray work
have DNA chip with small synthetic ssDNA sequences that are complimentary to genomic sequence with a SNP
DNA of interest is fragmented and separated into single strands then hybridized to the single stranded synthetic DNA on the slide
the empty position due to the DNP is revealed when the slide is washed and fluorescently labeled nucleotides are added
each color reflects the genotype of the individual at SNP. if homo- color will match one of the four, if hetero- spot will be a mix of two colors
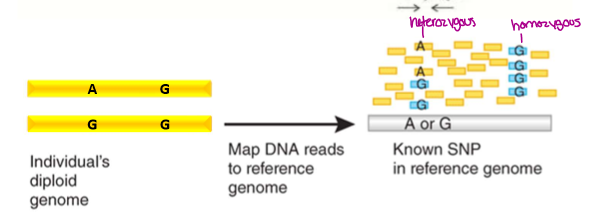
How does Genomic Resequencing work
Use illumina sequencing
sequence is compared to a reference genome and aligned based on similarities. If homo- all reads will be the same, if hetero- reads will be 50% one nucleotide and 50% another
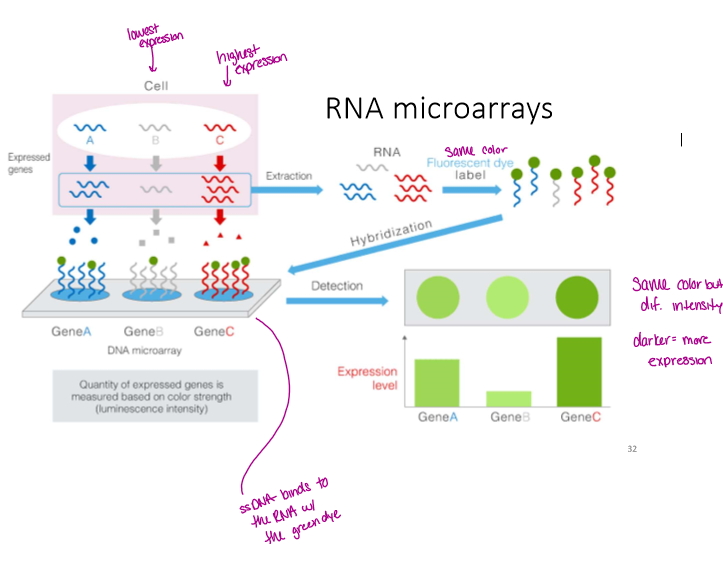
How does RNA microarray work
have a synthetic single stranded DNA bound to a slide
extract RNA from the sample of interest, it is reverse transcribed to make cDNA, add fluorescent label, then hybridize to the slide
highly expressed genes have RNA bound and are a bright color
can be used for competitive hybridization which shows relative expression of each gene between two samples. in this case you use two colors like red and green, a combo of the two is yellow, no expression is black
How does RNA sequencing work
collect mRNA from a tissue and reverse-transcribe it to make cDNA
sequence using illumina which results in reads which are then aligned to the species’ reference genome sequence
use read depth for each gene to assess level of expression and SNP if found in exons of expressed genes