D1.1: DNA Replication
1/40
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
41 Terms
Purpose of DNA Replication
Production of new, identical DNA to the existing DNA
DNA structure allows for repeated replication, without limitations to number of times
Replication Uses (2)
Reproduction - offspring need copies of parental DNA
Growth and tissue replacement - both require cell division and DNA must be replicated beforehand
What allows for high degree of accuracy during replication?
Semi-conservative model and complementary base pairing
Semi-Conservative Model
both original strands serve as templates for the new strands
-possible bc of copmlementary base pairng (A-T and C-G)
-produces DNA we/ one original (parent, template, antisense) strand and one new (daughter, nontemplate, sense) strand
Allows for high degree of accuracy during replication
Parent Strand
The template for constructing the new DNA double helix, AKA the antisense strand
Daughter Strand
the newly made stand in DNA replication
Complementary Base Pairing
Only complementary bases can be added to new strand b/c H-Bonds cannot form between non-complementary strands
*this is how DNA polymerase III recognizes and replaces mispairings*
Directionality of DNA Polymerase
All nucleotides have 2 covalent bonds, except for those at the ends of the strand
add new nucleotides to the 3' end
phosphate groups are at 5' end
**build 5'-->3' and read 3'-->5'
Deoxynucleoside Triphosphate
dNTPs
free floating dNTPs have 3 phosphates, DNA polymerase cleaves 2 phosphates off and uses the energy to form a phosphodiester bond at 3' end of strand
Significance of 3' End
DNA polymerase NEEDS a 3' end to attach dNTPs
Therefore it can extend, but CANNOT initiate replication (needs RNA primer)
Enzymes in DNA Replication (5)
Helicase, DNA Primase, DNA Polymerase III, DNA Polymerase I, DNA Ligase
Helicase
Separates/unwinds DNA by breaking hydrogen bonds btwn nitrogenous bases
This occurs at the origin of replication, creates 2 replication forks (one on either side)
two parental strands act as templates
DNA Primase
lays down short RNA primers (10-15 bp) which provide a 3' end for DNA polymerase III to bind to and add nucleotides
DNA Polymerase III
binds to template strand at 3' end of RNA primer, NEEDS A 3' END, CAN EXTEND BUT CANNOT INITIATE
Covalently bonds complementary DNA nucleotides, growing chain in 5'-->3' direction
Forms phosphodiester bonds between deoxyribose sugar of one nucleotide and phosphate of another
Cleaves two phosphates off dNTPs and uses energy to create bonds
PROOFREADS EACH NUCLEOTIDE
DNA Polymerase I
Functions as an exonuclease and polymerase
Removes RNA primers and replaces them with DNA nucleotides in 5'-->3' direction
*Cannot make 3' to 5' links, leaves a gap where sugar-phosphate bond would be*
Exonuclease
an enzyme that removes successive nucleotides from the end of a polynucleotide molecule
DNA Ligase
Covalently bonds sugar-phosphate backbone to form a continuous strand
JOINS 3' end to 5' of next nucleotide (the gap that DNA Polymerase I can't form bonds
Joins okazaki fragments
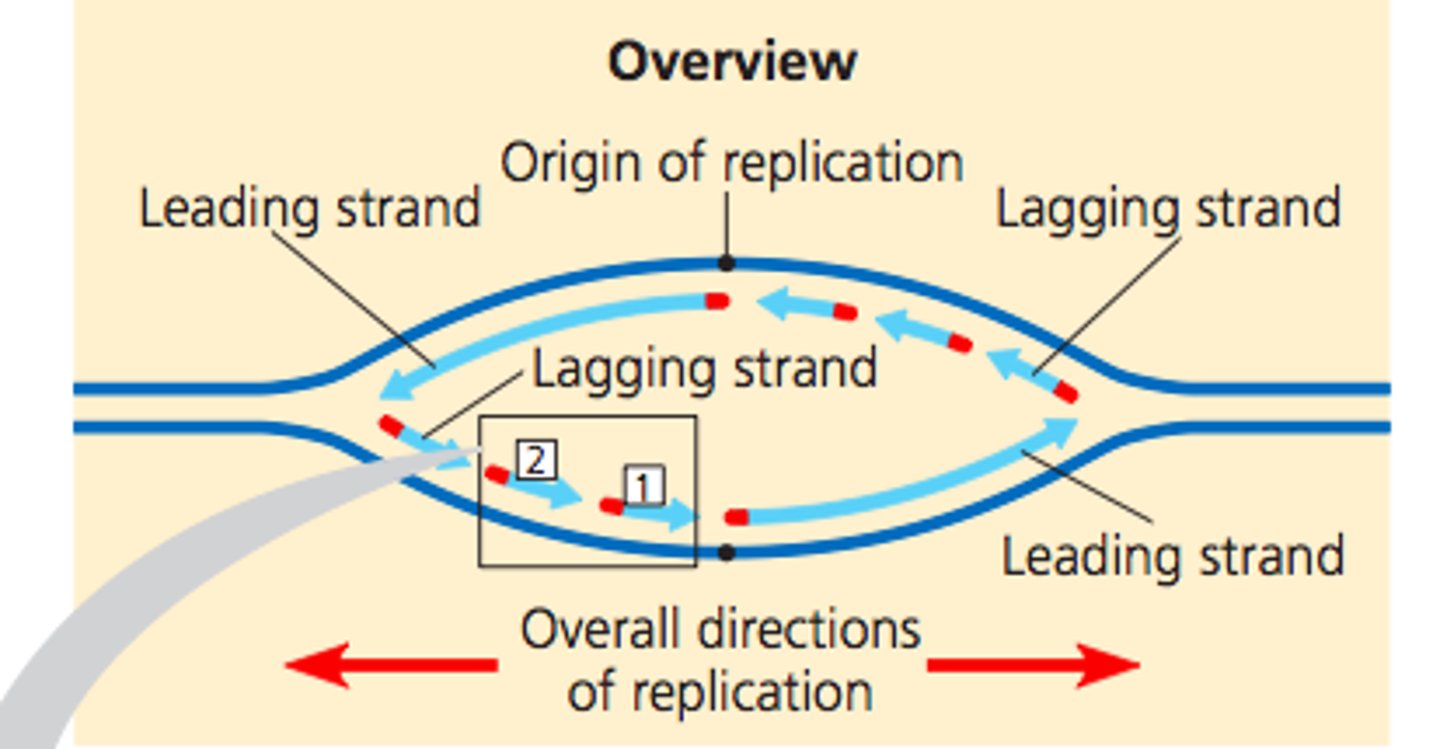
Leading Strand
Makes DNA continuously, grows TOWARDS replication fork (5'-->3')
Completed more quickly, needs RNA primer only ONCE
Lagging Strand
Makes DNA discontinuously, grows away from replication fork
*Has to backtrack to add nucleotides sequentially as replication fork moves away and exposes more of template strand*
Produces short DNA fragments (okazaki fragments), needs RNA primers every 100-200 bps
Okazaki Fragments
Small fragments of DNA produced on the lagging strand during DNA replication, joined later by DNA ligase to form a complete strand.
Leading vs Lagging Strand
Leading: elongate continuously into the widening replication fork
Lagging: replicates away from the fork, must wait until it widens to polymerize and is discontinuous, leading to Okazaki fragments
DNA Proofreading
Occurs immediately after wrong base has been added
Once recognized, DNA Polymerase III excises (removes) incorrect base, moves back one nucleotide and inserts correct base
Polymerase Chain Reaction (PCR)
Automated method of DNA replication, generates large amounts of specific DNA in short period of time, requires small quantity initially
*doubles about every 2-3 mins*
PCR Materials
1. DNA of interest (about 100 bps)
2. DNA primers
3. free nucleosides (dNTPs)
4. Taq polymerse
Taq polymerse
derived from a hot spring bacteria (Thermus aquaticus)
Can function in high temp. w/out denaturing
PCR Process
1. Denature
2. Anneal
3. Elongate
Denature
1st step in PCR at about 95 C, separates DNA strands by breaking H-bonds
Anneal
2nd step in PCR at about 55 C, allows primers to bind
Elongate
3rd step in PCR at about 72 C, optimal temperature for Taq polymerse, assembles new DNA strands
Gel Electrophoresis
Separates DNA fragments according to length
-place samples in "wells" in agar gel
-negatively charged phosphates of DNA travel towards ANODE (positive electrode of power source)
Shorter strands have less resistance to gel and therefore travel farther
Differing sizes separate into bands as they travel at different speeds
Dye in Gel Electrophoresis
Gel is treated w/ dye to make bands (DNA) visible
Banding Pattern
Determined by DNA sequence and type of restriction enzyme used
Size of DNA fragment is determined by comparing it to a know marker (ladder)
Restriction Enzyme
Enzyme that cuts DNA at a specific sequence of nucleotides
Satellite DNA
non-coding regions or "junk DNA"
Contains short tandem repeats (STRs) of DNA that are repeated a varied number of times (ex. TAGA or AGAA)
Short Tandem Repeats
Short sequences of DNA (2-7 bp) repeated a varied number of times
People vary in number and location, producing unique DNA profile (banding pattern)
13 or more STRs are used in DNA profiling (12 different STR locations in chromosome(
Excised (cut) using restriction enzymes and separated via Gel Electrophoresis
What are Gel Electrophoresis bands made of?
Each STR is cut into pieces (ex. for STR: AGAA --> atcgatcgatcgA BREAK GAAatcgatcgatcg)
This cuts the junk DNA into pieces w/ the STRs dividing them, and therefore different sections of junk DNA are different lengths bc STRs located randomly within Junk DNA
The different lengths of these junk DNA sections result in the banding pattern
**thickness of bands kinda irrelevant**
DNA Profiling Procedure
1. Collect DNA (From blood/semen)
2. amplify via PCR
3. cut all samples using same restriction enzyme (different STRS will generate different fragment lengths
4. Separate via Gel Electrophoresis
5. Compare profiles
DNA Profiling Uses (2)
Forensic Investigation and Paternity Testing
Forensic Investigation
to convict, suspect's DNA needs to have a 100% match w/ the DNA from crime scene
Population size in area is considered when determining the uniqueness of profile (ex. smaller population = more unique profiles = less STRs used)
Paternity Testing
children inherit their chromosomes from both parents
All the child's DNA must match either that of mother or father
*to find father, must be the man w/ banding pattern that matches everything that mom doesn't cover w/ her banding pattern for the kid*
Reason for Paternity Testing (2)
Men denying paternity to avoid financial responsibility
Children proving paternity for inheritance rights