elsaid - mechanisms of carcinogenesis and overview of anticancer drugs
1/47
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
48 Terms
neoplasia
new growth or autonomous growth of tissue
neopalsm
the lesion resulting form the neopalsia
benign
lesions characterized by expansive growth, frequently exhibiting slow rates of proliferation that do not invade surrounding tissues
malignant
lesions demonstrating invasive growth, capable of metastases to other tissues and organs
metastasis
secondary growths derived from a primary malignant neoplasm
cancer
malignant neoplasm
carcinogen
a physical or chemical agent that causes or induces neoplasia
genotoxic
carcinogn that interacts with DNA resulting in mutation
nongenotoxic
carcinogen that modify gene expression but do NOT damage DNA
carcinogenesis process:
initiation
carcinogenic agent (chemicals, radiation, viruses) causes DNA damage and cell mutation —> mutated cell
DNA modification
mutation
genotoxic damage
one cell division is necessary to lock in mutation
nonreversible
carcinogenesis process:
promotion
activation of oncogenes (proteins that help the cell proliferation) by promoter agent
no direct DNA modification
nongenotoxic
no direct mutation
multiple cell divisions necessary
clonal expansion of the initiated cell population
increase in cell proliferation and/or resistance to cell apoptosis
reversible
carcinogenesis process:
progression
malignant tumor caused by over expression of oncogenes
irreversible
changes from preneoplasia to neoplasia
mutation, chromosome rearrangement, DNA modification
original hallmarks of cancer
sustained proliferative signaling
cancer cells always want to proliferate (S —> M phase)
evading growth suppressors
cancer cells avoid signals from growth suppressor proteins
including angiogenesis
making new blood vessels to gain more nutrients and oxygen for growth
activating invasion and metastasis
different between different cancers (some can be more metastatic than others)
resisting cell death
enabling replicative immortality
enabling factors of cancer
avoiding immune destruction
express misfolded proteins (immunogenic antigens) on surface to evade immune system
deregulating cellular energetics
emerging hallmarks of cancer
tumor promoting inflammation
genome instability and mutation
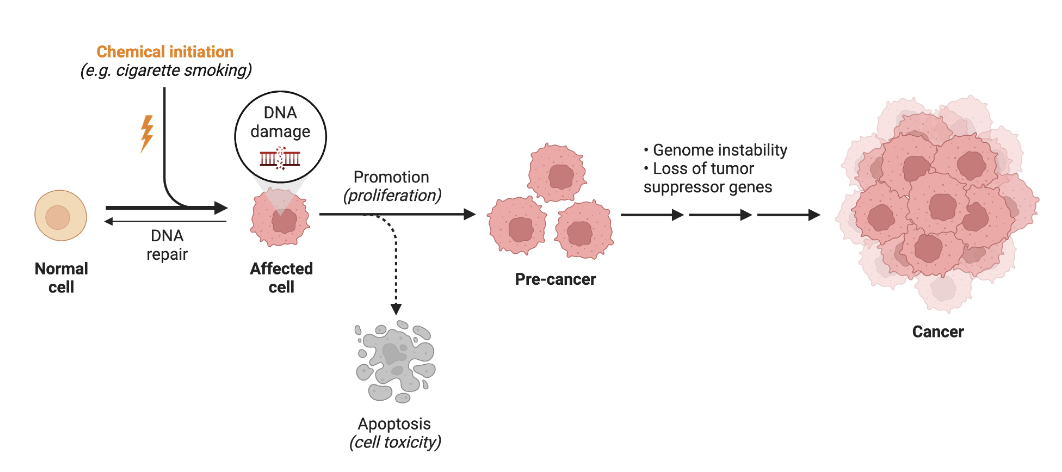
chemical carcinogenesis
chemical initiation causes DNA damage —> mutated/affected cell
DNA damage is greater than DNA repair
muted cells proliferate more than p53 (tumor suppressors) can cause apoptosis of the mutated cells
pre-cancer —> progresses into cancer
spectrum of DNA damage
single strand and double strand breaks
ds breaks = more significant and more opportunities for repair to go wrong
apurinic or apyrimidinic (abasic) sites (depurination or depyrimidation)
missing a base (either purine or pyrimidine base)
DNA adduct formation and crosslinking
adduct formation = chemical moiety that reacts w/ purine or pyrimidine base (covalent bond)
crosslinking = chemical moiety reacts w/ bases on BOTH strands of DNA and joins them together
what can cause DNA damage?
UV, x-rays, gamma rays
chemicals (e.g. reactive oxygen and nitrogen species, environmental toxins)
antineoplastic drugs (DNA-directed cytotoxic agents)
chromosomal rearrangements:
deletion
nucleotide(s) removed from a chromosome
chromosomal rearrangements:
duplication
a segment of DNA is duplicated
chromosomal rearrangements:
insertion
extra nucleotide base(s) are added into the DNA sequence
chromosomal rearrangements:
inversion
rearrangement of a segment of a chromosome is flipped/reversed
chromosomal rearrangements:
translocation
part of one chromosome breaks off and attaches to another chromosome OR two chromosomes exchange pieces
outcomes of chromosomal rearrangements
loss of function mutation in tumor suppressor genes
gain of function mutation in oncogenes
creation of fusion genes resulting in expression of fusion proteins (proliferation enhancement)
oncogenes
genes for growth factor receptors
EGFR or erbb1 (codes for epidermal growth factor receptor)
HER2 or erbb2 (codes for a growth factor receptor)
genes for signaling cascade proteins
KRAS (codes for guanine nucleotide-proteins with GTPase activity)
genes for cytoplasmic kinases BCR-ABL (codes for non-receptor tyrosine kinase)
tumor suppressor genes
APC (colon/rectum carcinoma)
BRC1 (breast and ovarian) and BRCA2 (breast)
encode for DNA repair proteins
DPC4 (pancreatic)
INK4 (melanoma, lung, brain)
MADR2 (colon/rectum)
P53 (multiple cancers)
induces apoptosis
PTEN (brain, melanoma, prostate)
Rb (retinoblastoma, bladder, breast)
VHL (renal cell carcinoma)
cellular growth signals:
stimulatory pathways
neighboring cells release growth-stimulatory factors
stimulatory factor binds to receptors on cell surface
cytoplasmic relay proteins activates transcription factors
transcription factors produce proteins that trigger cell division
cellular growth signals:
inhibitory pathways
neighboring cells release growth-stimulatory factors
stimulatory factor binds to receptors on cell surface
cytoplasmic relay proteins activates transcription factors
transcription factors produce proteins that inhibit cell division
stimulatory abnormality
cell divides in the absence of external growth factors
inhibitory abnormality
relay molecule is lost —> cell divides when it should not because inhibitory signal fails to reach nucleus
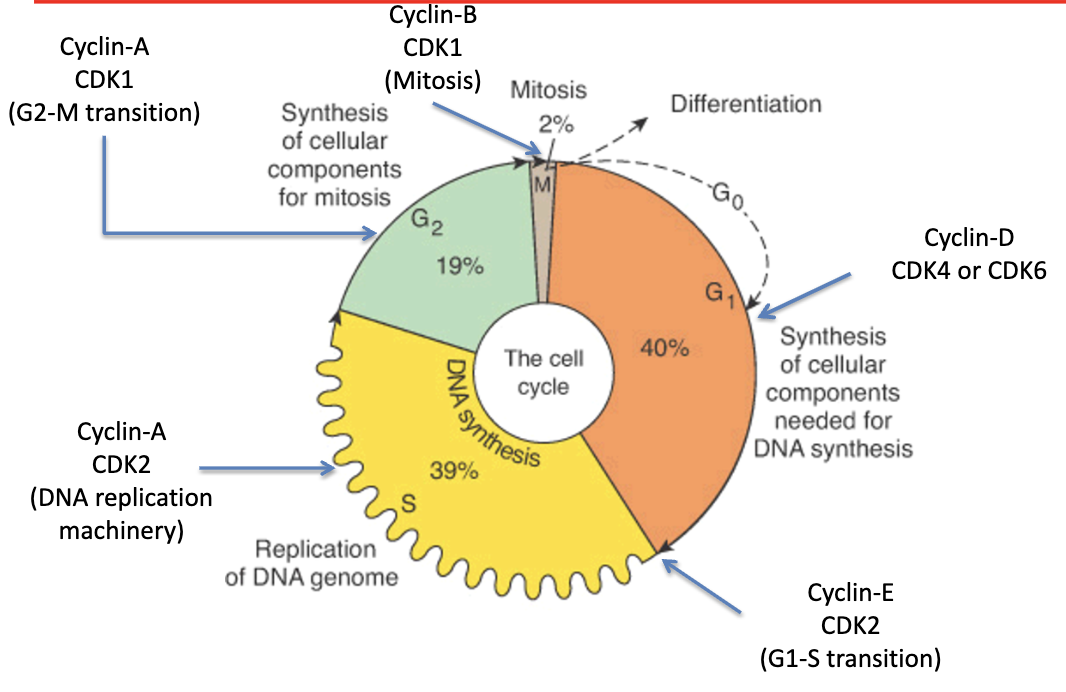
what regulates progression through the phases of the cell cycle?
a family of proteins called Cyclins and associated kinases (Cyclin-dependent Kinases; CDKs)
cyclin and CDK that controls cell cycle phase activity in G1 phase
Cyclin D1, D2, D3
CDK 4 and CDK 6
cyclin and CDK that controls G1/S phase transition
Cyclin E
CDK 2
cyclin and CDK that controls cell cycle phase activity in S phase
Cyclin A
CDK 2
cyclin and CDK that controls G2/M phase transition
Cyclin A
CDK 1
cyclin and CDK that controls mitosis
Cyclin B
CDK 1
cyclin and CDK that controls CAK, all cell cycle phases
Cyclin H
CDK 7
regulation of the cell cycle by Cyclin-Dependent Kinases (CDKs)
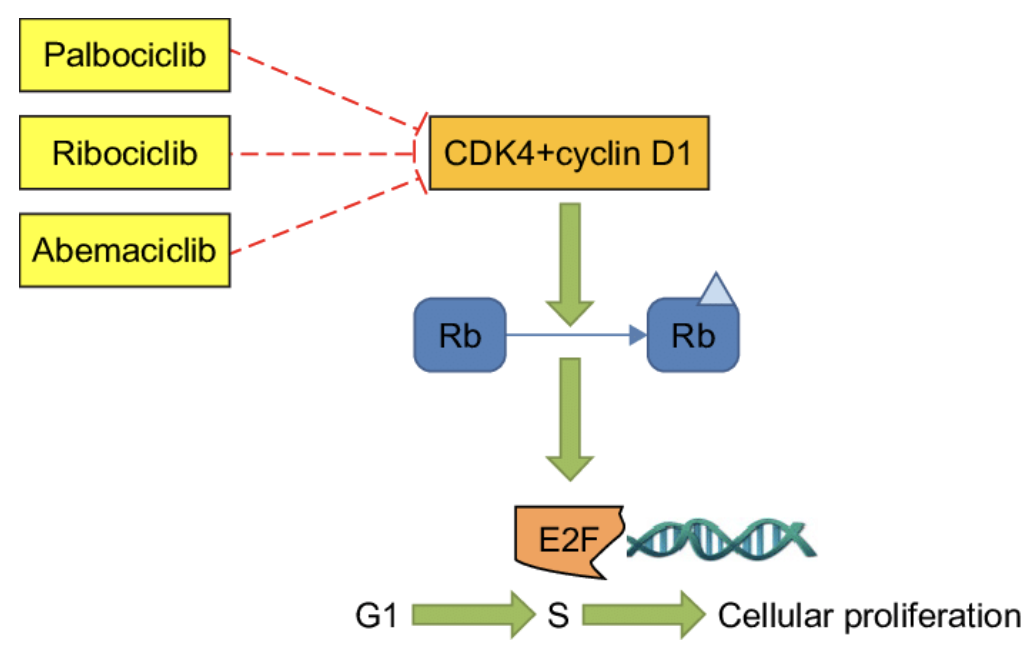
cell cycle inhibitors
palbociclib
ribociclib
abemaciclib
MOA of cell cycle inhibitors
inhibits CDK4 —> cells stuck in G1 and does NOT proliferate
CDK4 + cyclin D1 phosphorylates Rb
phosphorylated Rb activates E2F (elongation factor)
E2F promotes transition from G1 —> S —> cellular proliferation
conventional antineoplastic agents
alkylating agents
antimetabolites
antimitotic (antimicrotubules) agents
topoisomerase inhibitors
miscellaneous DNA directed agents
hormonal agents
SERMS (selective estrogen receptor modulators)
aromatase inhibitors
anti-estrogens
anti-androgens
inhibitors of steroidogenesis
immunotherapy
monoclonal antibodies
immune checkpoint inhibitors
therapeutic vaccines
targeted treatments
tyrosine kinase inhibitors
signal pathway inhibitors
differentiating agents
drugs that block topoisomerase function
camptothecins
etoposide
teniposide
daunorubicin
doxorubicin
drugs that forms adducts with DNA
platinum analogs
alkylating agents
mitomycin
cisplatin
temozolomide
mechanisms of resistance to the effects of anti-cancer drugs
up regulation of the MDR efflux pumps
decreased cellular uptake (if the anticancer drug requires a transporter for cellular uptake) by down regulation of uptake transporters
increased concentration of cellular target (enzyme, structural protein) or a mutated target (reducing binding affinity)
detoxification of the reactive species of the drug by glutathione
enhanced DNA repair and failure to induce apoptosis
distribution of anticancer agents
cancer mass is heterogeneous with cancer cells having different rates of proliferation depending on the presence of blood vessels and nutrient supply
hypoxic conditions can result in low cell proliferation and resistance towards anti-proliferative and cytotoxic agents
off-target effects of anticancer drugs account for their toxicities
EPR effect (enhanced penetration and retention):
molecules of certain size (liposomes, nanoparticles) accumulate more in cancer tissues than normal tissues