Chapter 10: Molecular Structure of Chromosomes
1/137
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
138 Terms
What are Chromosomes?
Structures that contain the genetic material
What complexes are Chromosomes?
Of DNA and Proteins
What is Genome?
Comprises all the genetic material that an organism possesses
The genome of bacteria is?
Typically a single circular chromosome
The genome of eukaryotes is?
Refers to one complete set of nuclear chromosomes
What other genome characteristics do Eukaryotes possess?
Possess a mitochondrial genome
What genome characteristic do plants have?
Have a chloroplast genome
What is the main function of genetic material?
To store the information required to produce the traits of an organism
How does genetic material store information required to produce the traits of an organism?
Accomplished via protein-encoding genes
What are four reasons DNA sequences are necessary?
Synthesis of RNA and cellular proteins
Replication of chromosomes
Proper segregation of chromosomes
Compaction of chromosomes (So they can fit within living cells)
Eukaryotic species contain?
One or more sets of chromosomes
What are the sets of chromosomes in Eukaryotic species composed of?
Each set is composed of several different linear chromosomes
Humans have 2 sets of 23 chromosomes
Each chromosomes contains?
A single, linear molecule of DNA
What are 2 characteristics of each chromosome?
Typically tens to hundreds of millions of base pairs
Typically a few hundred to several thousand genes
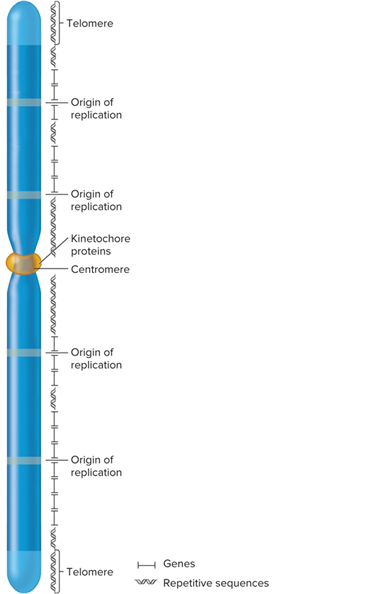
What does a Chromosome look like?
What is an example of simpler eukaryotes?
Yeast
What is the gene structure of yeast?
•Relatively short (e.g., several hundred bp long)
What are 2 examples of more complex eukaryotes?
Mammals
Flowering plants
What is the gene structure of more complex eukaryotes?
Genes are long
They tend to have many introns (noncoding intervening sequences)
Intron lengths from less than 100 to more than 10,000 bp
What are 3 types of DNA sequences are required for chromosomal replication and segregation?
Origins of Replication
Centromeres
Telomeres
What are Origins of Replication?
Chromosomal sites necessary to initiate DNA replication
Eukaryotic chromosomes contain many origins
What are Centromeres?
Regions that play a role in segregation of chromosomes
What are Telomeres?
Specialized regions at the ends of chromosomes
Important in replication and for stability
What are four key features of the Organization of Eukaryotic Chromosomes?
Eukaryotic chromosomes are usually linear.
Eukaryotic chromosomes occur in sets. Many species are diploid, which means that somatic cells contain 2 sets of chromosomes.
A typical chromosome is tens of millions to hundreds of millions of base pairs in length.
Genes are interspersed throughout the chromosomes. A typical chromosome contains between a few hundred and several thousand different genes.
What are the four chromosomal features of Organization of Eukaryotic Chromosomes?
Each chromosome contains many origins of replication that are interspersed about every 100,000 base pairs.
Each chromosome contains a centromere that forms a recognition site for the kinetochore proteins; required for chromosome sorting during mitosis and meiosis.
Telomeres contain specialized sequences located at both ends of the linear chromosome.
Repetitive sequences are commonly found near centromeric and telomeric regions, but they may also be interspersed throughout the chromosome.
The total amount of DNA in eukaryotic species is typically?
Much greater than that in bacterial cells
The total amount of DNA in eukaryotic species contains?
Many more genes
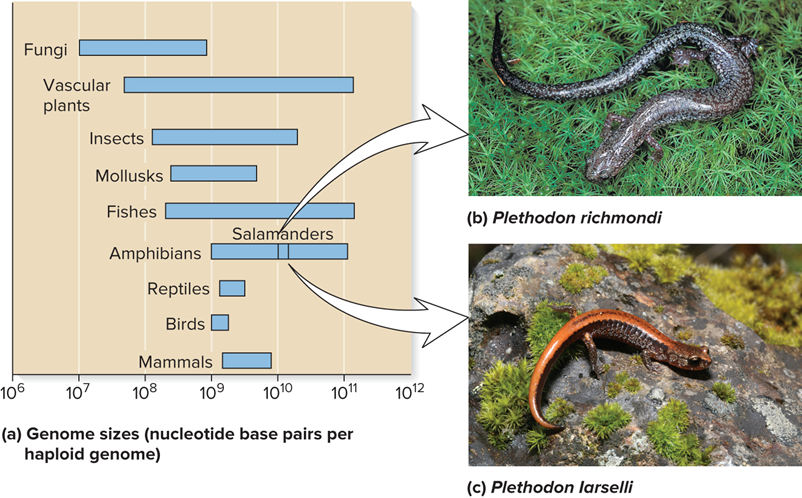
Eukaryotic genomes vary substantially in?
Size
The sizes for Eukaryotic Genomes in many cases?
This variation is not related to the complexity of the species
What are 2 examples of the variation in sizes for Eukaryotic Genomes that is not related to the complexity of the species?
For example, there is a two-fold difference in the size of the genome in two closely related salamander species
The difference in the size of the genome is not because of extra genes
Rather, the accumulation of repetitive DNA sequences
These do not encode proteins
What do Genome sizes for nucleotide base pairs per haploid genome, Plethodon richmondi and Plethodon Iarselli look like?
What is Sequency complexity?
Refers to the number of times a particular base sequence appears in the genome
What are the 3 types of repetitive sequences?
Unique or non-repetitive
Moderately repetitive
Highly repetitive
What are Unique or non-repetitive sequences?
Found once or a few times in the genome
Includes protein-encoding genes as well as intergenic regions
In humans, make up roughly 41% of the genome
What are Moderately Repetitive Sequences?
Found a few hundred to several thousand times
What are examples of moderately repetitive sequences?
Genes for rRNA and histones
Sequences that regulate gene expression and translation
Transposable elements
What are Highly repetitive sequences?
Found tens of thousands to millions of times
Each copy is relatively short (a few nucleotides to several hundred in length)
What are characteristics of Highly Repetitive Sequences?
Some sequences are interspersed throughout the genome
Other sequences are clustered together in tandem arrays
What is an example of Some sequences are interspersed throughout the genome?
Alu family in humans
Approximately 300 bp long
Represents 10% of human genome
Found every 5000-6000 bp
What is an example of Other sequences are clustered together in tandem arrays?
AATAT and AATATAT sequences in Drosophila
These are commonly found in the centromeric regions
What do Classes of DNA sequences look like?

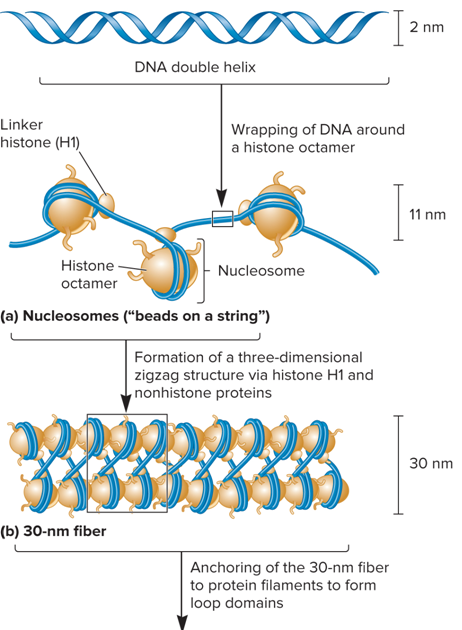
What is a nucleosome?
The repeating structural unit within eukaryotic chromatin
What are characteristics of Nucleosome?
Composed of a double-stranded segment of DNA wrapped around an octamer of histone proteins
A histone octamer is composed of two copies each of four different histone proteins
146 bp of DNA make 1.65 negative superhelical turns around the octamer
What does Nucleosomes showing core histone proteins look like?
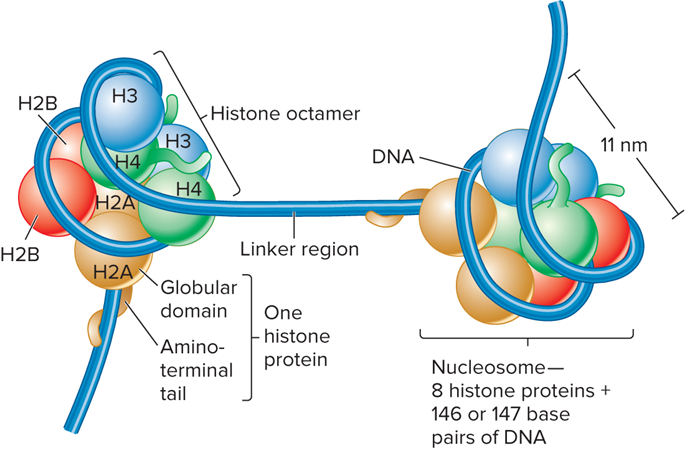
What are characteristics of Histone Proteins?
Basic
Contain many positively-charged amino acids (Lysine and Arginine)
These bind to the negatively-charged phosphates along the DNA backbone
Have a globular domain and a flexible, charged amino terminus or ‘tail’
What are the five types of Histones?
H2A
H2B
H3
H4
H1
Which histones are core histones?
H2A
H2B
H3
H4
How many of each core histone make up the octamer?
Two of each
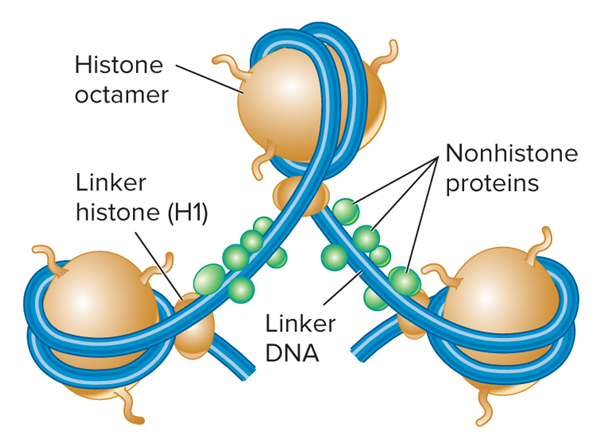
Which histone is called the linker histone?
H1
Why is H1 a linker histone?
Binds to DNA in the linker region
Less tightly bound to DNA than core histones
Helps to organize adjacent nucleosomes
What does the Molecular model for nucleosome structure look like?
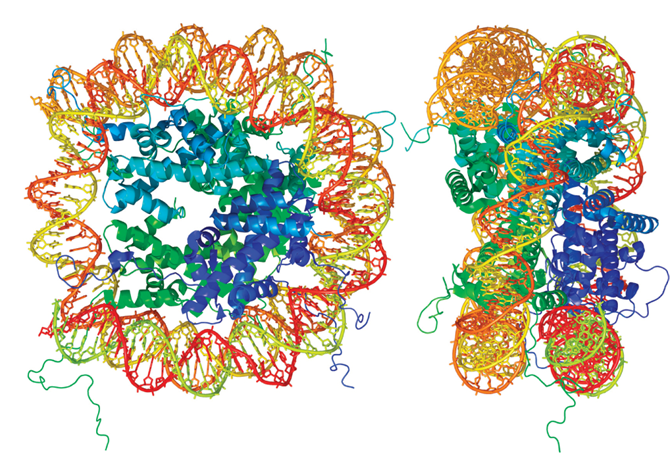
What do nucleosomes showing linker histones and nonhistone proteins look like?
Who proposed in 1745 the model of nucleosome structure?
Roger Kornberg
What did Kornberg base his proposal on?
Various observations about chromatin
What were the 3 observations about chromatin that Kornberg made?
Biochemical experiments
X-ray diffraction studies
Electron microscopy images
Who tested Kornberg’s Model?
Markus Noll
How did Markus Noll test Kornberg’s model?
Digest DNA with the enzyme DNase I
Accurately measure the molecular mass of the resulting DNA fragments using gel electrophoresis
The rationale is that the linker DNA is more accessible to DNase I than the DNA bound to core histones (Thus, the cuts made by DNase I should occur in the linker DNA)
What is Noll’s experiment test?
The beads-on-a-string model of chromatin structure
If Noll’s model was correct, DNase 1 should?
Preferentially cut DNA in the linker region
Thereby producing DNA pieces that are about 200 bp long
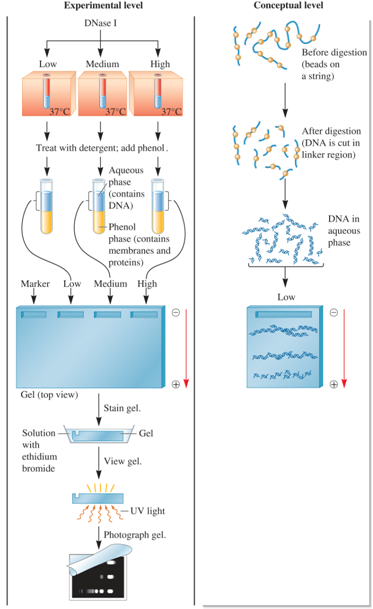
What was the process of Noll’s Experiment?
Incubate the nuclei with low, medium, and high concentrations of DNase I. The conceptual level illustrates a low DNase I concentration.
Isolate the DNA. This involves dissolving the nuclear membrane with detergent and treating the sample with organic solvent phenol.
Load the DNA into a well of an agarose gel and run the gel to separate the DNA pieces according to size. On this gel, also load DNA fragments of known molecular mass (marker lane).
Visualize the DNA fragments by staining the DNA with ethidium bromide, a dye that binds to DNA and is fluorescent when excited by UV light.
What did Noll’s Experiment look like?
What was the data of Noll’s Experiment
Note: The marker lane is omitted from this drawing.
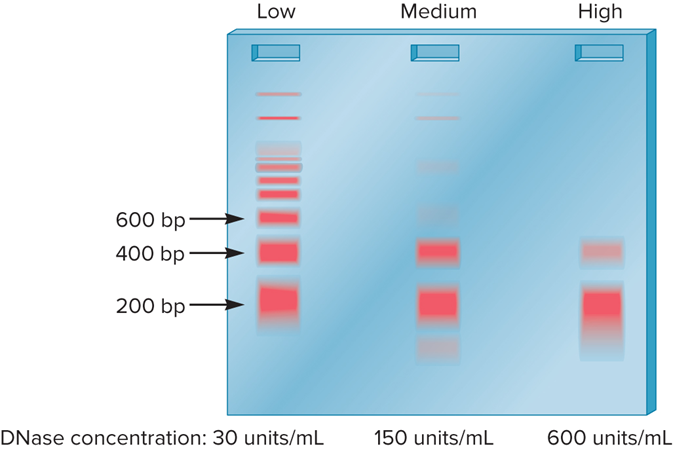
In Noll’s Experiment, At low or medium concentrations?
DNase I did not cut at every linker region
In Noll’s Experiment, At low or medium DNase I concentration?
Longer pieces were observed in multiples of 200 bp
In Noll’s Experiment, At high concentration?
All chromosomal DNA digested into fragments that are ~ 200 bp in length
Nucleosomes join to form?
A 30 nm Fiber
Nucleosomes associate with each other to form?
A more compact structure termed the 30 nm fiber
What plays a role in the compaction of Nucleosomes?
Histone H1
How does histone H1 play a role in the compaction of nucleosome structure?
At moderate salt concentrations, H1 is removed (The result is the classic beads-on-a-string morphology)
At low salt concentrations, H1 remains bound (Beads associate together into a more compact morphology)
What does H1 histone not bound-beads on a string look like?

What does H1 histone bound to linker region-nucleosomes more compact look like?
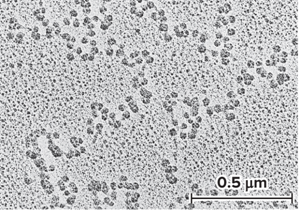
What does the 30 nm fiber shorten?
The total length of DNA another seven-fold
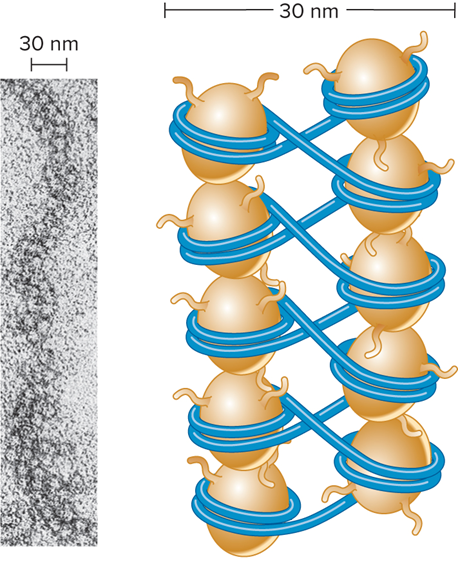
The 30 nm fiber structure has proven difficult to determine?
The DNA conformation may be substantially altered when extracted from living cells
Zigzag model has been proposed as a model for the structure of the 30 nm fiber
What does the micrograph of a 30 nm fiber and the zigzag model look like?
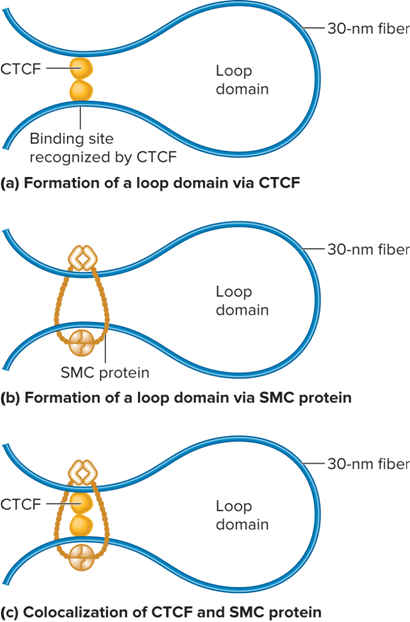
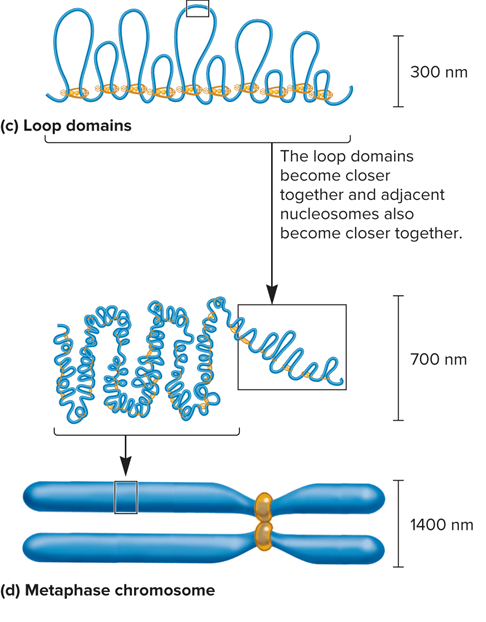
What is Third level of compaction?
30nm fiber folds into loop domains
What is CCCTC binding factor (CTCF)?
Binds to 3 regularly spaced repeats of the sequence CCCTC
How is a loop formed?
Two different CTCFs bind to the DNA and then bind to each other to form a loop
A second mechanism of loop formation is carried out by SMC proteins
An SMC protein forms a dimer that can wrap itself around two DNA segments and form a loop
In addition, SMCs may wrap around sites that are bound by CTCF dimers
What does the formation of a loop look like?
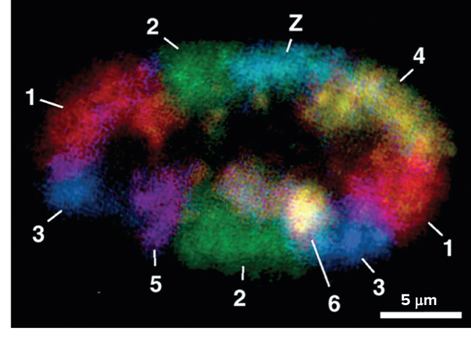
Each chromosome in the cell nucleus can be found in?
A discrete chromosome territory
How is each chromosome in the cell nucleus known to be location in a discrete chromosome territory?
In the studies by Thomas and Christoph Cremer and others through fluorescent staining in which each chromosome is shown in a different color
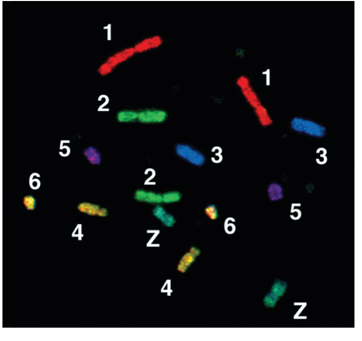
What do Metaphase Chromosomes look like?
What do Chromosomes in the cell nucleus during interphase look like?
The compaction level of interphase chromosomes is not?
Completely Uniform
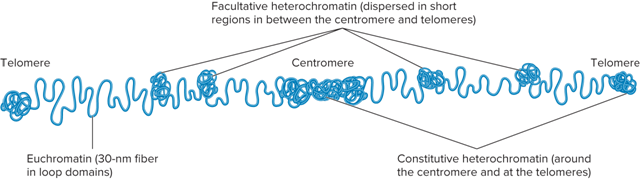
What is Heterochromatin?
Tightly compacted regions of chromosomes
Transcriptionally inactive (in general)
Loop domains compacted even further
What is Euchromatin?
Less condensed regions of chromosomes
Transcriptionally active
The 30 nm fiber forms loop domains
What are the 2 types of Heterochromatin?
Constitutive heterochromatin
Facultative heterochromatin
What is Constitutive Heterochromatin?
Regions that are always heterochromatic
Permanently inactive with regard to transcription
Usually contain highly repetitive sequences
What is Facilitative Heterochromatin?
Regions that can interconvert between euchromatin and heterochromatin
What does Constitutive/Facultative heterochromatin look like?
How does the structure of eukaryotic chromosomes change during cell division?
As cells enter M phase, the level of compaction changes dramatically
By the end of prophase, sister chromatids are entirely condensed
Two parallel chromatids have an overall diameter of 1,400 nm and are much shorter than in interphase
These highly condensed metaphase chromosomes undergo little gene transcription
What does Figure 10.23 (a. Nucleosomes (“beads on a string” and b. 30 nm fiber) look like?
What does Figure 10.23 (c. loop domains and d. metaphase chromosome) look like?
What are four characteristics of Metaphase Chromosomes?
Controversy regarding whether or not nonhistone proteins form a scaffold to organize metaphase chromosomes
When a metaphase chromosome is treated with salt to remove histones, loops are still attached to a darkly staining scaffold
Scaffold appears to contain SMC proteins
However some researchers are not convinced a protein scaffold plays a major role in metaphase chromosome organization
What does a Metaphase chromosome look like?
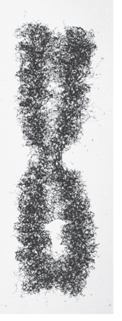
What does Metaphase chromosome treated with high salt to remove histone proteins look like?
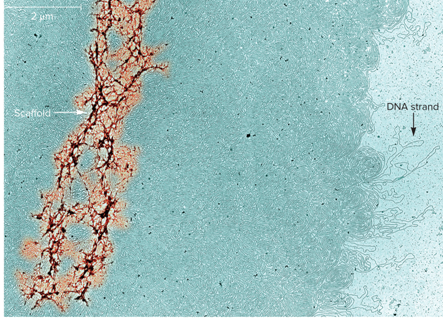
Condensin and Cohesin promote?
The Formation of Metaphase Chromosomes
Two multiprotein complexes help to form and organize?
Metaphase chromosomes
What are 2 examples of two multiprotein complexes?
Condensin
Cohesin
What is Condensin?
Plays a critical role in chromosome condensation
What is Cohesin?
Plays a critical role in sister chromatid alignment