13. Analyzing Mixtures of DNA, RNA and protein
1/68
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
69 Terms
nucleic acid hybridization
means for detecting complementary in DNA or RNA samples
Southern blotting
DNA detection
northern blotting
RNA detectionn
protein-protein interaction
means detecting proteins with other proteins (antibodies)
western blotting
protein detection
what do we need in order to detect the specific gene or specific product of gene expression “target”
DNA/RNA need a labeled hybridization probe = 10 mer - 100’s mer of nucleotides complementary gene of interest (DNA; Southern) or mRNA (northern)
protein: need a labeled antibody
TARGET is unknown sequence or protein and PROBE is
known sequence or antibody
what does it mean to be labeled
something identifiable, detectable, measurable
Hybridization Probe or Antibody
use radioactive atoms in nucleotides (typically “label” T for DNA, U for RNA probes)
use covalently attached fluorescent molecules, variety of enzyme conjugates
incorporated into the probe during synthesis of probe
method for detection (measurement) of radioactivity, or light or enzymatic product
labeled probe will
hybridize with target and be detected
Source of our labeled probe DNA
whole or partial coding sequence for the gene of interest from same organism or
whole or partial coding sequence for the gene of interest from a different organism, or
sequence of the gene from the same gene family as our gene of interest
in general: we use labeled nucleotides to perform
de novo DNA or RNA sequences from one of the above DNA sources as a template
homologous probe
probe and target sequences are perfect match (100% complementarity)
heterologous probe
when probe and target are not 1—% complementary, some degree of mismatching of bases
Source of our labeled probe for protein
“target” protein in sufficient quantities for antibody production (monoclonal, polyclonal)
typically, labeling with enzymes or fluorescent tags can be added after antibody production and purification
how complementary is our probe with the target
it depends ont he template used to make the probe
percentage of sequence identity (complementarity, homology) between target sequence and probe determines the hybridization conditions
higher homology
allows higher stringency
The conditions under which you hybridize (temperature, salt concentration) change
the “minimum” homology that will be required for hybridization to be successful between target and probe
controlling these conditions is controlling stringency
stringency considerations are also affected by
probe size and the actual sequence used as a probe or targeted (e.g repeats in either)
the ultimate goal while performing nucleic acid hybridization (for both DNA and RNA detection)
is to bind the probe to the target in a controlled manner
depending on the source of the probe sequence the probe could have
more or fewer mismatches with the target nucleic acid when they hybridize
percentage of identity (similarity) between the target and the probe determines STRINGENCY which is
a measure of the tolerance for mismatches between the two
higher similarity - higher stringency
less changes for mistakes/mismatch
lower similarity - lower stringency
higher chance for mistakes/mismatch
homologous probe (100% similarity)
heterologous probe (<100%)
what do we need in order to detect the specific gene or specific product of gene expression “target”
labeled probe (nucleic acid or protein)
sour e of our labeled probe?
same organism, related not related
how complementary is our probe with the target
stringency and sequence similarity
southern blotting detects
specific DNA fragments (identify specific restriction fragments in a complex mixture of fragments)
southern blotting can be used for
estimating the # and position of gene copies in a genome
restriction mapping of genomic fragments
detection of
cloned sequences
transgenes
homologous sequences in different genomes
repetitive sequences
southern blotting steps
digest DNA with restriction endonucleases
perform agarose gel electrophoresis on the DNA fragments from different digests
DNA fragments fractionated by size (visible under UV light if gel is soaked in ethidium bromide)
soak gel in NaOH: neutralize
transfer (blot) gel to nitrocellulose or nylon membrane using Southern blot technique
DNA fragments are bound to the membrane in positions identical to those on the gel
hybridize membrane with radioactively labeled probe
expose membrane to X-ray film; resulting autoradiograph shows hybridized DNA fragments
DNA in each and every cell of the organism is the same; however,
the set of synthesized proteins is different in different tissue/cell types , and/or during the life cycle
Temporal control
genes that are expressed at a precise time during the life cycle of an organism, this is also called developmental regulation. (e.g ovalbumin, globin in haemoglobin, plant seed storage proteins)
spatial control
genes expressed in a specific tissue or cell type. also described as tissue-specific expression. (e.g different genes expressed in liver cells, muscle cells or root tip cells)
many genes are both temporally and spatially controlled such that they are expressed in
a specific tissue at a precise stage of development of the tissue (e.g developing seed or flower expresses certain genes specific to those organs at a precise stage of development)
induced gene expression
change in types or amount of gene expression in response to environmental signals, exposure to chemical subtance or physiological stress (e.g thermal stress - heat shock genes; gene expression controlled by steroid hormones; toxic substances such as heavy metals, antibiotics or anaesthetics)
northern blotting
mRNA is transcribed from the protein coding DNA - first in gene expression
total RNA (all three major groups) is isolated from cells and electrophoresed
Normal blotting detects specific mRNAs
northern blotting is similar procedure to southern blotting but
no denaturation (RNA is already ss)
Northern blotting method can be used to determine steady-state level of a specific transcript in a certain RNA mixture =
abundance of specific mRNA at certain time, under certain conditions
northern blotting depends on
both transcription and degradation rate for that specific mRNA
we could compare abundance of mRNA isolated from
different tissues of one organism (e.g brain and muscle)
same tissue from different organism (brain from frog and brain from bird)
different treatments or conditions (time studies, normal versus transformed/treated)
remember probe is designed to detect certain mRNA, the one which is
transcribed from our gene of interest which codes for our protein of interest
in situ hybridization: probe binds to
complementary nucleic acids within cell or tissue (proves are the same as for southern and northern)
in situ hybridization: similar to southern and northern blotting BUT identifies
genes directly in chromosomes (FISH - Fluorescence In Situ Hybridization shown)
transcripts (mRNA) directly in cell or tissue for developmental expression studies, following treatments or environmental changes
expression studies of more htan one gene at the same time
many genomes have been sequenced - info on various gene sequences easily obtainable
pattern of genes expressed in a cell is characteristic of its present state
all or most differences in a cell state are correlated with changes in mRNA levels of genes
even expression patterns of uncharacterized genes may provide clues to their function
traditional methods are “one gene in one experiment” (obtained by northern hybridization” do not
show the whole picture of total gene expression in a cell and interaction of gene products
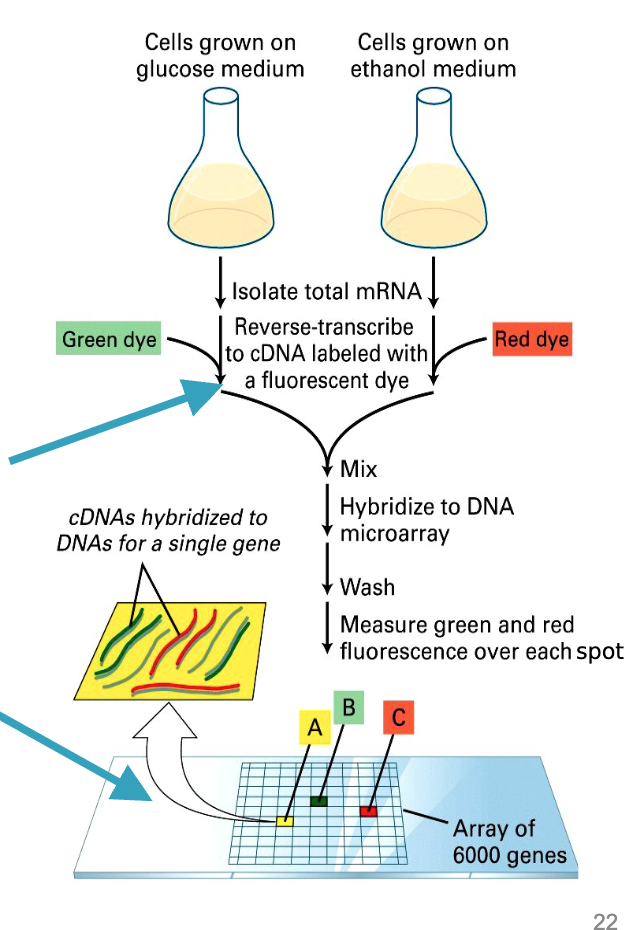
yeast genome microarray
target (unknown sequences) are getting labeled during experiment (= labeled cDNAs; made from isolated mRNAs during experiment)
Probe is known (made by/for us); its fixed and unlabeled
DNA microarray summary Grow cells in two conditions (e.g., glucose vs. ethanol).
Isolate mRNA → reverse transcribe to cDNA with different fluorescent dyes (green vs. red).
Mix cDNAs → hybridize to microarray (each spot = 1 gene).
Scan colors:
Green → higher in condition 1
Red → higher in condition 2
Yellow → equal in both
Principle: Complementary base pairing to measure relative gene expression.
measuring gene expression by measuring translation
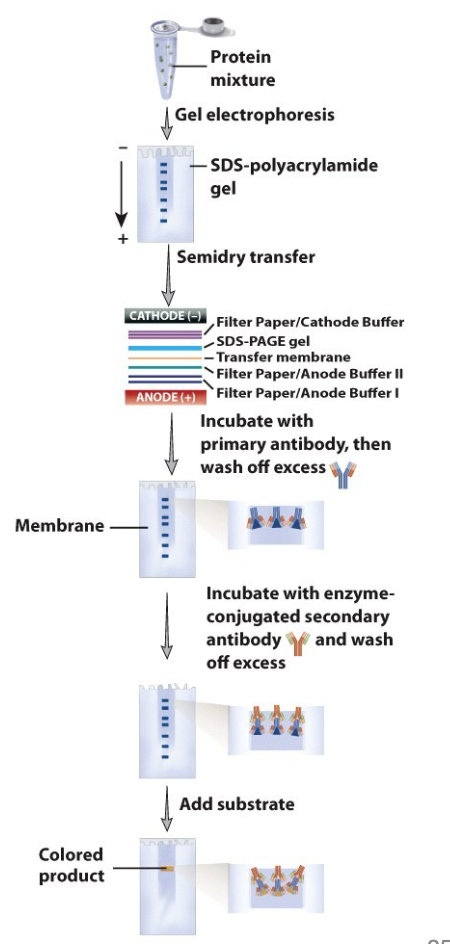
SDS sodium dodecyl sulfate polyacrylamide gel electrophoresis and Western Blotting
SDS - negatively charged detergent; binds to hydrophobic protein regions
helps protein unfolding
protein bind lots of SDS in constant ratio to their mass
gives them a negative charge proportional to their mass (equal charge density per unit length)
protein intrinsic charge is masked
electrophoresis under denaturing conditions:
proteins migrate towards the positive electrode when voltage is appleid to a gel
separate on the basis of molecular weight rather than intrinsic charge
can add a 2nd dimension (isolectric point) = 2D gel
proteins detected in the gel by different stains: varying sensitivity and specificity
western blotting
SDS PAGE - proteins are denatured by heat and detergent (SDS) and electrophoresed, separating by size
the proteins are transferred to a membrane (here using “electroblotting”) - western blotting
the membrane is incubated with an antibody specific for one of the proteins
bound antibody is detected by a secondary antibody that is conjugated to an enzyme or tagged (radioactive or fluorescent tags) = visualization
mapping transcriptional start sites
DNA regulatory elements (promoter elements) that control transcription are often located near the start site of transcription of the gene
in order to define the promotor region for a gene it is necessary to know the start of transcription
also: important regulatory elements in the 5’ UTR of mRNA
methods used to locate transcriptional start sites
S1 nuclease protection (S1 mapping)
primer extension
S1 nuclease protection (S1 mapping)
collect RNA from study organism, hybridize to labelled antisense probe, digest remaining single stranded nucleic acids (unhybridized organism RNA and probe). run on gels for analysis
primer extension
hybridize selected mRNA with radiolabeled 20-50 mer complimentary to a region close to 3’ end. reverse transcript to produce cDNA then compare to original DNA sequence for transcript start site
gene expression
process in which information carried by a gene is converted into observable product
transcription
first step in gene expression - process where one strand of a DNA molecule is used as a template for synthesis of a complementary RNA, mRNA which carries information for a specific protien
one-cell system (bacteria)
has to survive and reproduce
gene expression regulation - to adjust to changes in its nutritional environment to enable cell growth and cell division
multi-cellular organism: has to survive and reproduce but also to grow and develop; different parts of the body - different function
gene expression regulation during:
development (time)
tissue differentiation (space and '‘space/time combo”)
stress (as a response to environmental stress - induction)
regulation of gene expression: prokaryotes
transcription initiation is controlled
transcription and translation occur in the same compartment
mRNA is polycistronic, without introns and has a short half-life
regulation of gene expression: eukaryotes
eukaryotic cell is “compartmentalized” - regulation in each compartment
gene expression can be regulated at various levels in eukaryotes
transcription initiation is most important level fo regulation
transcription (general) similar process to DNA synthesis
enzyme - multi-subunit complex produces nucleotide strand in 5’ to 3’
mg2+ is a co-factor (necessary to add to buffers for in vitro transcription)
both DNA strands could be templates for RNA synthesis
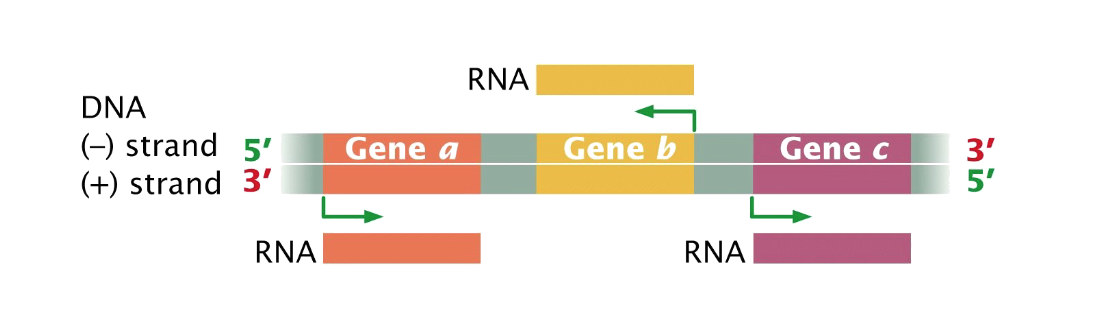
enzyme is RNA polymerase (RNAP)
RMAP does not require a primer
Promoter sequences (RNAP binding sites) are
asymmetrical RNAP is positioned so it can only transcribe one strand from one promoter
DNA is unwound locally only
ATP is not required (DNA and RNAP undergo spontaneous reversible structural changes - energetically favourable state)
Product is single stranded RNA
released from the template immediately (DNA helix re-forms) many copies from same gene
precursors use ribose -
ribonucleotide triphosphates (rNTPs)
RNAP is less
efficient in proofreading (mistake every 10^4 nucleotides)