bacteria (completed)
1/44
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
45 Terms
![<p>structure of bacteria [10]</p>](https://knowt-user-attachments.s3.amazonaws.com/7fc4bb49-5067-4a03-85a0-e018fdec8cbd.jpg)
structure of bacteria [10]
bacteria chromosome
plasmid
starch granule
70s ribosome
cytoplasm
cell membrane
peptidoglycan cell wall
capsule
fimbria
flagellum
bacteria chromosome structure and where its found [3]
ds circular DNA form looped domains by associating with DNA binding proteins
then supercooling form highly condensed DNA
found in nucleoid region
how does the F plasmid differ from the bacteria chromosome [2]
F plasmid has significantly fewer base pairs
F plasmid contains non-essential genes such those that confer advantages like anti-biotic resistance unlike bacteria chromosome which codes for essential genes like enzymes for cell metabolism
what is the structure and role of the F plasmid [5]
small circular autonomously replicating DNA molecule
F factor on the F plasmid codes for proteins necessary for the formation of sex piles and subsequently the mating bridge
to allow conjugation to occur between bacteria
allow bacteria genes to be transferred between bacteria increase genetic variation between bacteria
containing genes which confer advantages such as antibiotic resistance
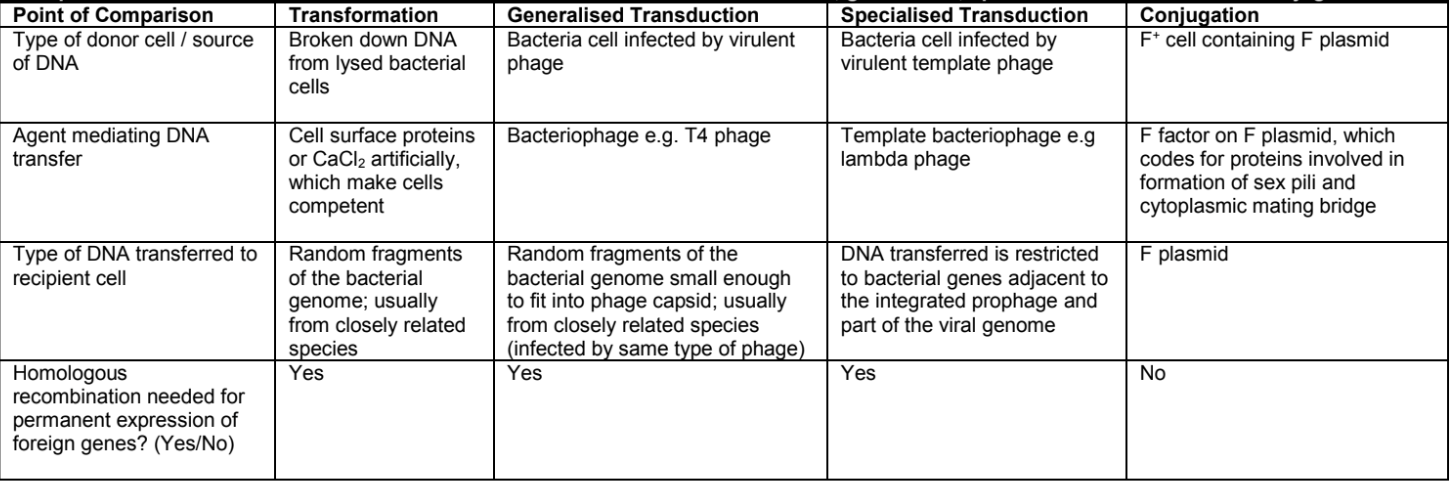
outline the process of bacteria conjugation [3]
sex plus of F bacterial cell makes contact with F- cell and retracts bringing F- cell closer for mating bridge to be formed between both cells
one of the 2 strands of plasmid DNA is nicked and transferred from the F+ cell to the F- cell through the mating bridge via the rolling circle mechanism and the other DNA strand is used as a template for elongation
the single stranded F plasmid DNA circularised in the F- cell and is used as a template to synthesise a complementary strand for the ds F plasmid FNA resulting in a F+ cell
what methods give rise to genetic variation in bacteria genomes [3]
conjugation
transformation
transduction (general or specialised)
describe the rolling circle DNA replication process [5]
one strand of the ds circular DNA is nicked by nuclease, breaking the phosphodiester bond
the 3’OH end elongated by DNA polymerase using intact stand as a template
newly synthesised strand displaces the 5’ end of the nicked strand and transferred to the recipient bacterium via the mating bridge
upon completion of 1 unit length of the plasmid, another nick occurs to release the original strand which recircularises
for the synthesis of complementary DNA strand to form ds F plasmid
outline the process of bacteria transformation [3]
fragments of foreign naked DNA from lysed bacteria cells in the surrounding medium are taken up by bacteria cells by cell surface proteins
foreign DNA is incorporated into bacteria chromosome via homologous recombination (crossing over at homologous regions)
if the foreign DNA contains a different allele that is now expressed in the bacteria cel, the bacteria cell has transformed
outline the process of general transduction [4]
phage infects a bacterium, injecting viral genome into host cell
bacterial DNA degraded into small fragments, which may be randomly packaged into a capsid head during spontaneous assembly of new viruses
upon cell lysis, defective phage infects another bacterium and inject bacteria DNA from the previous hose cell into new bacterium
foreign DNA can replace homologous regions in recipient cell chromosome if homologous recombination occurs which may express different allele from the previous host
outline the process of specialised transduction [5]
temperate phase infects a bacterium, injecting viral genome into the host cell
viral DNA is integrated into bacteria chromosome forming a prophage which mat be improperly excised to include adjacent segments of bacterial DNA during induction event
bacterial DNA may be packaged into a caps hear during spontaneous assembly of new viruses
(last 2 points same as general transduction)
upon cell lysis, defective phage infects another bacterium and inject bacteria DNA from the previous hose cell into new bacterium
foreign DNA can replace homologous regions in recipient cell chromosome if homologous recombination occurs which may express different allele from the previous host
benefits of conjugation for the recipient bacteria [2]
gain new alleles that when expressed allow it too survive in a different environment, antibiotic resistance
use of new resources like metabolites
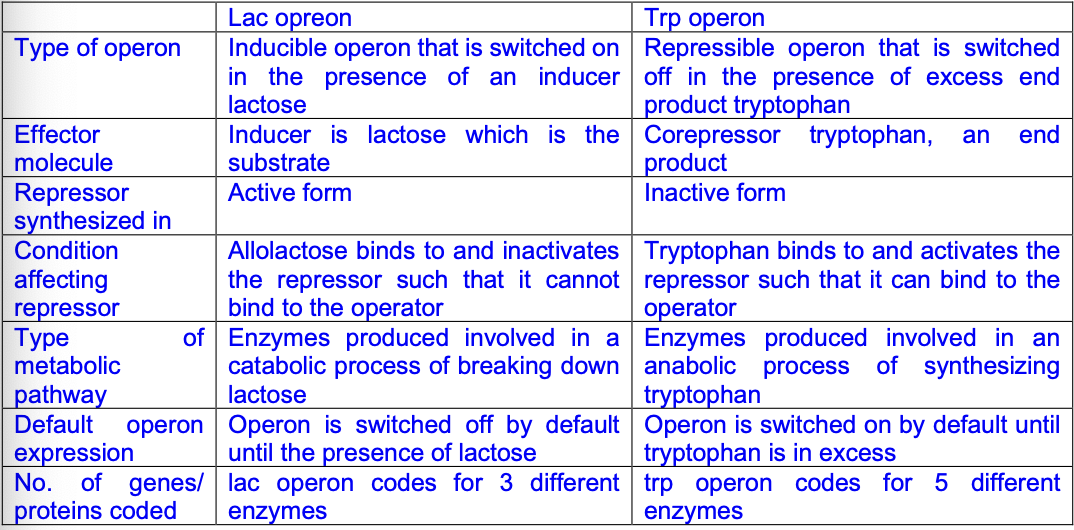
difference between pathways for repressible operons and inducible operons [2]
repressible is associated with anabolic while inducible is associated with catabolic pathways
state the preferred respiratory substrate [1[]
glucose over lactose
define operon [2]
a cluster of functionally related genes under the control of the same promoter and operator
allowing functionally related proteins synthesised together via translation of polycistronic mRNA
define promoter [4]
GTF and RNA polymerase binding site for the formation of TIC to initiate transcription
has critical elements
TATA BOX (precise location of transcription start site where GTF binds)
CAAT and GC BOX (improve efficiency of promoter)
define operator [1]
repressor protein binding site, prevent RNA polymerase from binding to the promoter, prevent transcription
define polycistronic mRNA [2]
messanger RNA containinig base sequences coding for amino acids for several proteins
contain many start (AUG) and stop (UGA, UAA, UAG) codons depending on number of polypeptides (3 sets for lac, 5 sets for trp)
define structural genes [1]
gene that codes for protein product the forms part of a structure of has an enzymatic function in a metabolic pathway
define regulatory genes + examples [3]
codes for protein involved in regulating expression of structural genes
not found within operon but diffusible operator to exert its effect
CAP and repressor
define effector and 2 examples [3]
a small molecule that binds to a specific protein, causing a conformation change and this regulating biological activity of the protein
allolactose in lac operon (inducer)
tryptophan in trp operon (corepressor)
advantages of operons [4]
allow functionally related proteins synthesised at the same time
economical use of energy and resources, expressing genes only when needed
allow bacteria respond rapidly and appropriately to changes in the environment
give them selective advantage over those who cant
![<p>Lac operon structure [3SG, 1 RG, 2P, 1O, I] INDUCIBLE OPERON</p>](https://knowt-user-attachments.s3.amazonaws.com/1fb03880-4b22-408d-99cc-d310e4154032.png)
Lac operon structure [3SG, 1 RG, 2P, 1O, I] INDUCIBLE OPERON
structural genes:
lac Z → beta-galactosidase (hydrolyse lactose to 1. glucose and galactose / 2. allolactose)
lac Y → permease (facilitate movement of lactose into the cell)
lac A → transacetylase
regulatory gene:
lac L → lac repressor (bind to DNA binding site of operator and allosteric site of allolactose)
promoter:
RNA polymerase binding site
Catabolite activator protein (CAP) binding site
operator: (overlap with promoter)
lac repressor binding site
inducer:
allolactose
![<p>Trp operon structure [5SG, 1RG, P, 1O,C] REPRESSIBLE OPERON</p>](https://knowt-user-attachments.s3.amazonaws.com/432db034-611e-4802-8a8e-0326c0bb531e.png)
Trp operon structure [5SG, 1RG, P, 1O,C] REPRESSIBLE OPERON
structural genes:
trp E, D, C, B, A → code for enzyme in tryptophan biosynthesis pathway
regulatory gene:
trp R → trp repressor (bind to DNA binding site of operator and allosteric site of tryptophan)
promoter:
RNA polymerase binding site
operator: (overlap with promoter)
trp repressor binding site
corepressor
tryptophan
negative regulation [1]
lac operon turned OFF by repressor protein in the absence of lactose
positive regulation
lac operon turned ON by CAP protein in the presence of glucose
lac operon (NO lactose and glucose) - just talk abt lactose
OFF
regulatory gene lacl constitutively transcribed to produce lac repressor protein
lac repressor protein in active form binds to the operator via the DNA binding site
in the absence of lactose, repressor protein remains active binds to the operator prevent RNA polymerase from accessing the promoter
preventing the transcription of structural genes
lac operon (YES lactose NO glucose)
ON
presence of lactose, a few molecules of lactose enter the cell with the help of permease
lactose converted to allolactose by beta galactosideas
allolactose (inducer) binds to the allosteric site of the repressors, repressor becomes inactive as the DNA binding site of the repressor conformation altered and cannot bind to the DNA binding site of the operator as its tertiary structure altered and no longer complementary
RNA polymerase can access and bind to the promter to initiate transcription of structural genes
structural genes transcribed as a single polycistronic mRNA coding for 3 different proteins to be translated together
absence of glucose, cAMP levels increase
cAMP bind to the allosteric site the CAP forming CAP- cAMP complex, activating CAP which binds to the CAP binding site within the promoter
this increase the affinity of promoter region of RNA polymerase, increase frequency of transcription of lac operon structural genes
lac operon (NO lactose YES glucose)
OFF
regulatory gene lacl constitutively transcribed to produce lac repressor protein
lac repressor protein in active form binds to the operator via the DNA binding site
in the absence of lactose, repressor protein remains active binds to the operator prevent RNA polymerase from accessing the promoter
preventing the transcription of structural genes
presence of glucose, cAMP levels low
catabolite activator protein (CAP) not activated as few cAMP bound to CAP
hence no CAP binding to CAP binding site in the promoted and no up regulation of transcription of lac operon
lac operon (YES lactose and glucose)
OFF
presence of lactose, a few molecules of lactose enter the cell with the help of permease
lactose converted to allolactose by beta galactosideas
allolactose (inducer) binds to the allosteric site of the repressors, repressor becomes inactive as the DNA binding site of the repressor conformation altered and cannot bind to the DNA binding site of the operator as its tertiary structure altered and no longer complementary
RNA polymerase can access and bind to the promter to initiate transcription of structural genes
structural genes transcribed as a single polycistronic mRNA coding for 3 different proteins to be translated together
presence of glucose, cAMP levels low
catabolite activator protein (CAP) not activated as few cAMP bound to CAP
hence no CAP binding to CAP binding site in the promoted and no up regulation of transcription of lac operon
to memorise about glucose
U glucose → D cAMP
D glucose → U cAMP
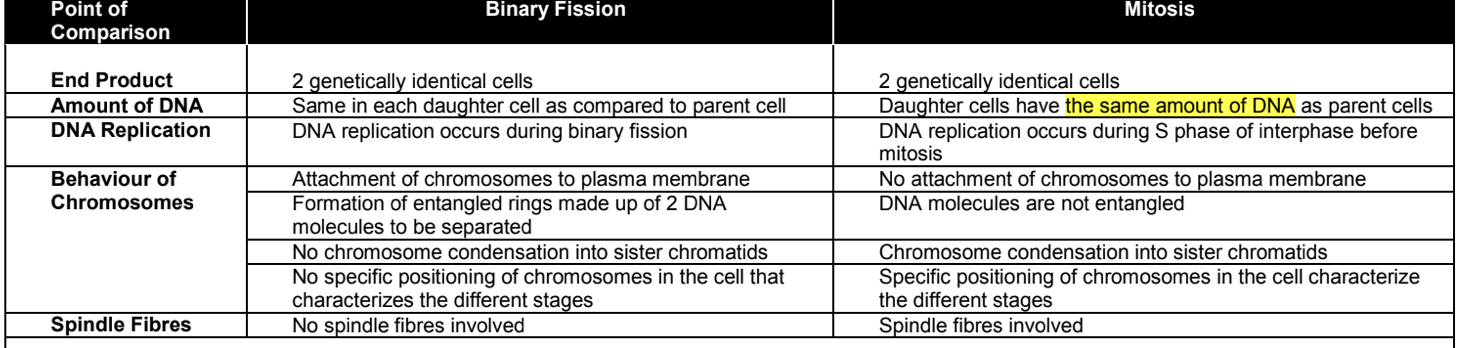
what happens to the gnome of bacteria during binary fission [5ed]
bacteria genome made upon of DNA replicates by semi-conservative replication
DNA unzipped by helices by breaking the hydrogen bonds between the bases of 2 strands
each parental strand serve as a template for synthesis of daughter strand
free deoxyribonucleotides CBP with bases on parental strand
DNA polymerase catalyse formation of phosphodiester bonds between adjacent nucleotides
describe hoe binary fission produces genetically identical bacteria [6]
DNA replication begins at the original of replication DNA unzipped by breaking hydrogen between base pairs of 2 strands forming a replication bubble
DNA replicate by semiconservative replication each original strand used as template for synthesis of complementary daughter strand by CBP
2 newly formed ori move to opp poles of the cell and attach to plasma membrane
cell elongate to prepare for division
DNA circular with no free ends and 2 daughter molecules interlocked with completion of replication
enzyme topoisomerase cut, separate and reseal 2 DNA molecules
invagination of plasma membrane and the deposition of new cell wall eventually divide parent cell into 2 daughter cells
COMPARISON: binary fission vs mitosis
COMPARISON: transformation vs general transduction vs specialised transduction vs conjugation
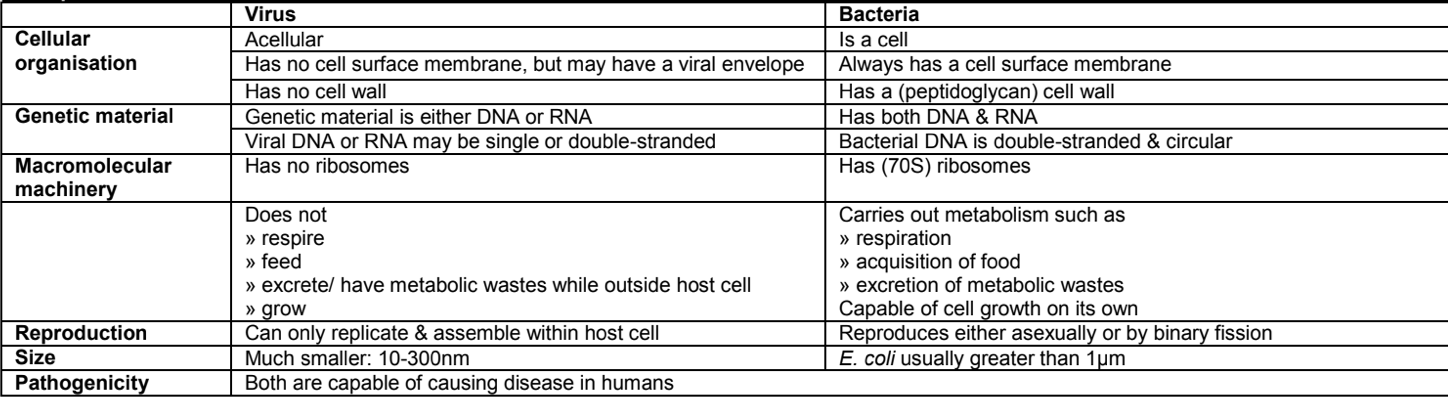
COMPARISON: viruses vs bacteria
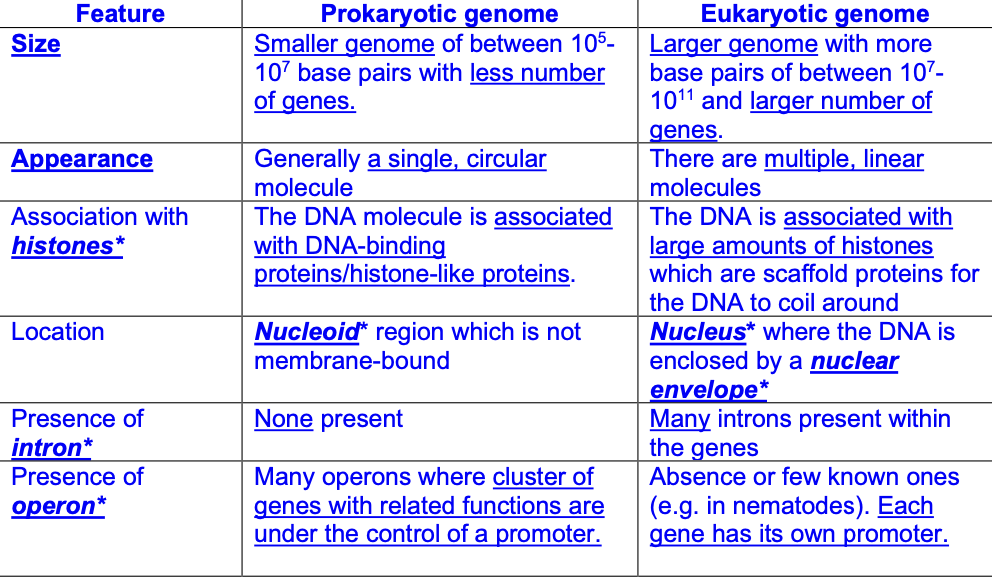
COMPARISON: pro genome vs euk genome [6]
effect of a mutation of a single structural gene and on the other structural genes [2]
truncated of non-functional protein coded for the mutated structural gene
the other proteins still produced as they are translated indecently as they each have their own start and stop codon on the polycistronic mRNA
describe mutation of lac l gene and its effect [5]
mutation of the gene results in a different mRNA transcribed and thus different amino acids translated, defective lac repressor
causing a conformation change DNA binding site of the repressor, bind reversibly to the operator
also causes a change in the allosteric site of the repressor, cannot bind to the inducer (allolactose) to become inactivated, remain bounded to the operator
lac operon permanently switched off
no structural genes transcribed thus levels of lactose etc same
why is it lactose not broken down immediately even though glucose is absent [1]
time is needed for the transcription of lac Z and subsequent translation to produce beta-galactosidase for lactose to be broken down
why regulatory gene located far away but can still regulate operon expression [1]
the repressor protein coded by regulatory gene can diffuse to the location of the operator to bind and exert its effect
trp operon (high tryptophan) [4]
trp repressor protein synthesised in inactive form
as tryptophan accumulates, it acts as a corepressor and binds to the allosteric site of the repressor, changing its conformation to be active
active repressor bind to operator, prevent binding of RNA polymerase to the promoter
prevent expression of operon
trp operon (low tryptophan) [2]
trp repressor protein synthesised in active form and unable to bind to the operator
RNA polymerase can bind to the promoted and allow expression of operon
COMPARISON: how DNA arranged in pro vs euk genome
euk:
DNA coils around histone proteins to form nucleosome
follow by coiling around itself to form solenoids
supercoiling of the loops consider into metaphase chromosome
pro:
associated with relatively fewer proteins (histone-like proteins) to form loops
super coiling cause further compaction
how are lac and trp operon similar [4]
both consists of several genes under control of single promoter
both have operator region for binding of active repressor to prevent transcription
genes coding for functionally related proteins synthesised together as a unit
operon produces a polycistronic mRNA with several start and stop codons
COMPARISON: lac and trp operon